The untranslated regions (UTRs) of mRNA molecules regulate their stability and translation efficiency which influences protein expression levels. The regulatory elements found in both 5' UTR and 3' UTR regions of mRNA molecules directly influence translation efficiency and stability. Before the coding region of mRNA lies the 5' UTR which acts as a necessary element for initiating protein synthesis. Ribosome binding sites (RBS) and internal ribosome entry sites (IRES) function as critical translation initiation points and facilitate ribosome attachment while being situated within this mRNA region. Secondary structures like hairpins and stem-loops in the 5' UTR region affect how ribosomes bind to and scan mRNA molecules. The translation efficiency of mRNA depends on the structural features found within the 5' UTR because these structures demonstrate unique stability characteristics and spatial arrangements. The sequence positioned downstream of the stop codon in the 3' UTR controls both mRNA stability and lifespan. The regulatory elements present in the sequence control how fast mRNA molecules degrade. AU-rich elements located in the 3' UTR attract protein complexes which then initiate mRNA degradation. The mRNA stability increases when stem-loop structures in the 3' UTR protect the mRNA from exonuclease degradation. Protein binding sites in the 3' UTR have dual roles where they can enhance mRNA stability while also causing mRNA destabilization. Proteins influence mRNA stability through their interactions with sequence motifs and structural elements present in the 3' UTR. The human β-globin 3' UTR segment serves as a common element in IVT mRNA design because it boosts protein expression levels.
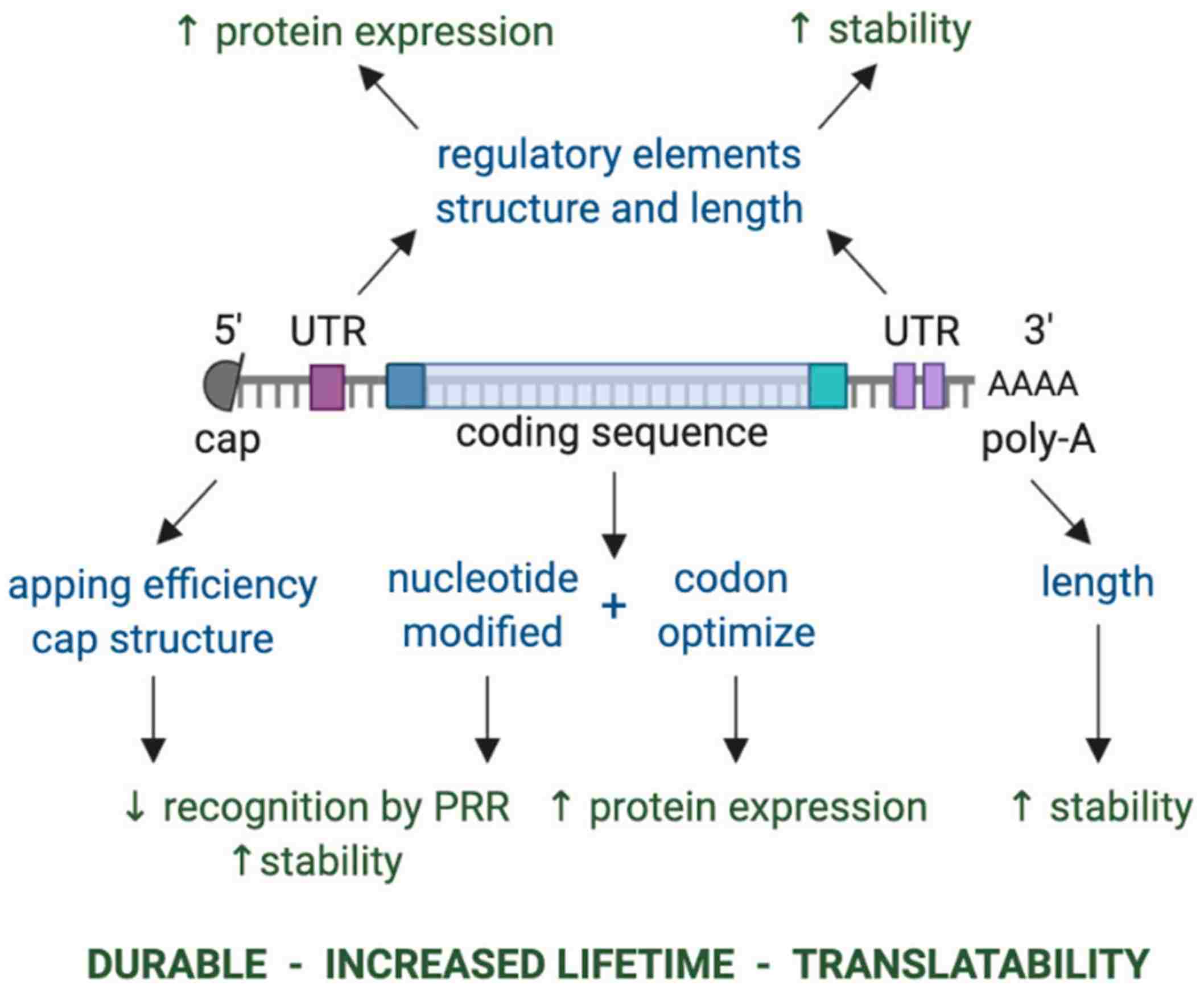 Fig. 1 Key components of in vitro transcribed mRNA that determine the level and duration of expression of the encoded protein.1
Fig. 1 Key components of in vitro transcribed mRNA that determine the level and duration of expression of the encoded protein.1
The Kozak sequence within the 5' UTR serves as a key determinant for the efficiency of translation initiation. The 5'-GCC(A/G)CCAUGG-3' sequence improves ribosome binding to AUG start codons to initiate translation. The nucleotides at positions -3 (preferably A or G) and +4 (G) are essential for achieving maximum translation efficiency near the AUG codon. Maximizing protein expression levels requires strategic Kozak sequence optimization which makes it vital for designing effective mRNA constructs. Achieving maximum translation efficiency requires precise alignment between the Kozak sequence design and the consensus sequence. A strong Kozak sequence shows significant benefits when applied to boost expression of inherently low-expressing genes and to synthesize therapeutic proteins. AI-driven computational tools enable enhanced design of Kozak sequences to achieve optimal gene expression in different host cell types. A neutral amino acid such as Val, Ala or Gly can be added following the start codon when the second amino acid in the sequence does not feature a start-G codon to include a G and enhance the Kozak sequence.
Secondary structures formed by hairpins and stem-loops in the 5' UTR prevent ribosome attachment and scanning resulting in reduced translation efficiency. The optimal performance of mRNA requires the reduction of secondary structures. RNA secondary structure prediction involves algorithms that decide the minimum free energy (MFE) state of RNA sequences. The algorithms enable mRNA sequence modification that reduces secondary structure strength without changing the amino acid sequence. The latest research introduced a new method for improving mRNA secondary structure optimization by increasing the nucleotide sequence's minimum free energy (MFE). The method integrates simulated annealing and linear regression to determine synonymous codon combinations that maximize MFE thus decreasing secondary structure strength. The suggested method leads to an increase of more than 40% in MFE which causes a significant reduction in the interference of secondary structures during translation.
The 3' UTR contains multiple regulatory motifs which critically influence mRNA stability and translation rates. RNA-binding proteins such as HuR recognize AREs as regulatory motifs to stabilize mRNA molecules. The microRNA response elements within mRNA sequences act as binding sites for microRNAs which govern both the degradation of mRNA and the translation process. Optimization of regulatory motifs results in improved mRNA stability and enhanced translation efficiency. The addition of AU-rich elements to the mRNA 3' UTR activates intracellular HuR to protect and stabilize the RNA leading to enhanced RNA stability and improved translation.
To maintain mRNA stability and extend its half-life researchers must remove the degradation signals found in the 3' UTR. Some sequence motifs along with structural elements serve as degradation signals that help cellular machinery detect and quickly degrade mRNA molecules. Elements that contain high levels of adenine and thymine in the 3' UTR are thought to control mRNA stability and the rate of mRNA degradation. Removing degradation signals from the 3' UTR will result in a significant improvement in mRNA stability. To enhance mRNA stability researchers can remove sequences that draw RNA-degrading enzymes and change their structure or they can incorporate elements that protect mRNA from degradation.
The subsection titled 'In Silico Screening and Validation of 5' UTRs' introduces the high-throughput pipelines developed for in silico identification of 5' UTRs usable in mRNA therapies.
Researchers have developed high-throughput in silico screening pipelines to identify potential 5' UTRs for mRNA therapeutic applications. The UTR-Insight pipeline creates an all-encompassing endogenous 5' UTR database through the collection of natural 5'UTR sequences from various species such as primates, mice, and viruses. The database estimates minimum ribosome landing (MRL) points for sequences while filtering out sequences containing upstream start codons (uAUGs) and selecting testing candidates.
The study team selected 20 5' UTRs for experimental testing simultaneously with the establishment of the In silico group. The research team identified 10 high-translation-efficiency 5' UTR sequences that had been experimentally validated and previously reported through literature mining including the hHBA 5' UTR from the BioNTech COVID-19 mRNA vaccine (BNT162b2) as well as the hCYBA 5' UTR for use as controls. The study selected 20 5' UTR sequences with high translation efficiency from NGS data to function as comparison controls.
Scientists prepare multiple primers which target various 5' UTR sequences to carry out experimental validation. Reverse PCR creates plasmid vector templates through selective amplification of regions lacking the native 5' UTR from the vector template. Researchers confirm band size through agarose gel electrophoresis analysis of PCR products and simultaneously purify and quantify DNA templates.
Scientists perform polysome profiling MPRA on mRNA libraries with variable 5' UTR regions in order to determine how efficiently short variable 5' UTRs are translated. Scientists introduce these libraries into cells followed by lysate isolation to carry out polysome profiling and RNA extraction. The RNA produced is reverse transcribed and then experiences template switching followed by qPCR amplification before it is sequenced using Illumina technology.
During kinetics measurements scientists create a calibration line with spike-in controls to regularize read counts obtained from designed 5' UTRs. Protein production estimates derive from integrating mRNA abundance across multiple time points. MEME and related software tools enable researchers to perform motif enrichment analysis to discover sequence motifs that exhibit high translation efficiency.
At BOC Sciences, we offer specialized services for designing 5' and 3' untranslated regions (UTRs) that are precisely tailored to your mRNA application's translational efficiency, half-life, and expression kinetics. Whether you're working on vaccines, protein therapies, or gene editing tools, our UTR optimization strategies are data-driven and application-specific.
The 5' UTR plays a critical role in ribosome recruitment and initiation. Our services include:
The 3' UTR contributes to mRNA stability, localization, and translation duration. We provide:
We tailor UTRs based on the unique needs of each therapeutic modality:
We use advanced computational tools and wet-lab validation to ensure your UTRs perform as expected:
Secondary structure prediction (e.g., mFold, RNAfold)
Immunogenicity and motif screening
In vitro translation assays and reporter gene testing
Comparative expression analysis across UTR variants
Whether you need to optimize an existing mRNA construct or design a UTR architecture from scratch, our team is here to help you unlock greater translational efficiency and molecular stability.
Partner with us to streamline your path to high-performance mRNA therapeutics.
5'UTR sequences control ribosome binding and initiation, with optimized Kozak motifs and minimized secondary structures significantly enhancing translation efficiency for high protein expression.
We optimize regulatory motifs, remove degradation signals, and incorporate stabilizing elements like stem-loops to increase mRNA half-life and extend expression duration.
We tailor UTRs based on specific goals - rapid expression for vaccines, sustained output for therapeutic proteins, or precise timing for gene editing applications.
We use polysome profiling, massively parallel reporter assays (MPRA), and motif enrichment analyses to quantitatively assess translation efficiency and stability of UTR variants.
Our computational algorithms predict minimum free energy states and suggest synonymous changes to reduce inhibitory structures while maintaining coding sequence integrity.
High-throughput in silico screening efficiently identifies optimal UTR candidates from databases, enabling selection of sequences with superior translation and stability before experimental validation.
References