The capping process stands as a vital modification step for the therapeutic application of mRNA molecules. The 7-methylguanosine (m7G) 5′ cap structure connected through a 5′-5′ triphosphate bridge serves multiple essential functions including mRNA stability preservation and translation efficiency enhancement while enabling immune system evasion. The mRNA cap structure functions to protect the mRNA molecule against 5′ exonuclease degradation which maintains its structural integrity in the cytoplasm. Eukaryotic translation initiation factors (eIFs) together with eIF4E form a cap complex that directs ribosomes to mRNA molecules for translation. Specific interactions between ribosomes and mRNA enable the translation of mRNA which leads to efficient protein synthesis. Translation efficiency benefits from methyl groups at the 2′-O position of the first two nucleotides present in both cap 1 and cap 2 structures. The capping process functions as an essential step that allows mRNA molecules to exit the nucleus. The cap structure links to nuclear export components to move mRNA from the nucleus to the cytoplasm where translation occurs. This step directs the mRNA molecule to the cellular protein synthesis apparatus.
The m7G cap structure also called m7GpppN which serves as cap 0 emerges from the attachment of a m7G group to the first mRNA nucleotide via a 5′-5′ triphosphate linkage. The eukaryotic translation initiation factors including eIF4E bind to the mRNA structure which facilitates accurate translation. Cells treat Cap 0 mRNA as foreign RNA which activates an innate immune reaction upon its introduction. The mRNA structure known as Cap 1, represented by m7GpppNm, exhibits an extra methylation modification at the 2′-O position on the nucleotide that follows the m7G cap. The additional methylation enables the mRNA to escape recognition by the innate immune system which diminishes the chance of an immune response during in vivo administration. Research indicates that Cap 1 mRNA demonstrates greater stability and improved translation efficiency relative to Cap 0 mRNA. The predominant form of RNA in higher eukaryotes such as humans is Cap 1 which plays a critical role in differentiating self RNA from non-self RNA to reduce immunogenicity.
The presence of extra methylation in Cap 1 increases its interaction with translation initiation factors resulting in better translation efficiency than Cap 0. The ability of Cap 1 to avoid detection by the innate immune system makes it the optimal choice for therapeutic uses where reducing immune activation is essential. The additional methylation of Cap 1 mRNA enhances its stability by shielding it against degradation.
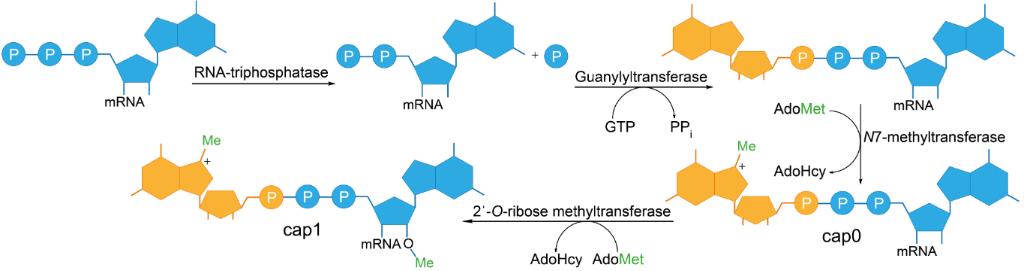 Fig. 1 Schematic representation of enzymatic 5′-cap formation in eukaryotic mRNA.1
Fig. 1 Schematic representation of enzymatic 5′-cap formation in eukaryotic mRNA.1
Chemical synthesis methods cannot currently produce mRNAs at full length which makes enzymatic methods essential for their preparation. During in vitro transcription (IVT) RNA polymerase facilitates the addition of ribonucleoside 5′-triphosphates (NTPs) to form an RNA chain that mirrors the nucleotide sequence of the DNA template. Scientists use RNA polymerases from T7, T3, or SP6 bacteriophages to start synthesis because these enzymes require only a single nucleotide to begin transcription. Each polymerase needs a specific DNA sequence known as the promoter site to attach to the template and begin IVT. RNA transcription using T7 class III promoters (such as ϕ6.5) results in RNA chains beginning with guanosine while initiation from T7 class II promoters (such as ϕ2.5) starts with adenosine. Initiating RNAs with pyrimidines is more challenging. RNA synthesis during IVT involves elongation of the oligoribonucleotide chain from 5′ to 3′ resulting in the production of 5′-triphosphorylated mRNA. Producing 5′-capped mRNA necessitates either extra steps for post-transcriptional capping or adjustments to the IVT protocol for co-transcriptional capping.
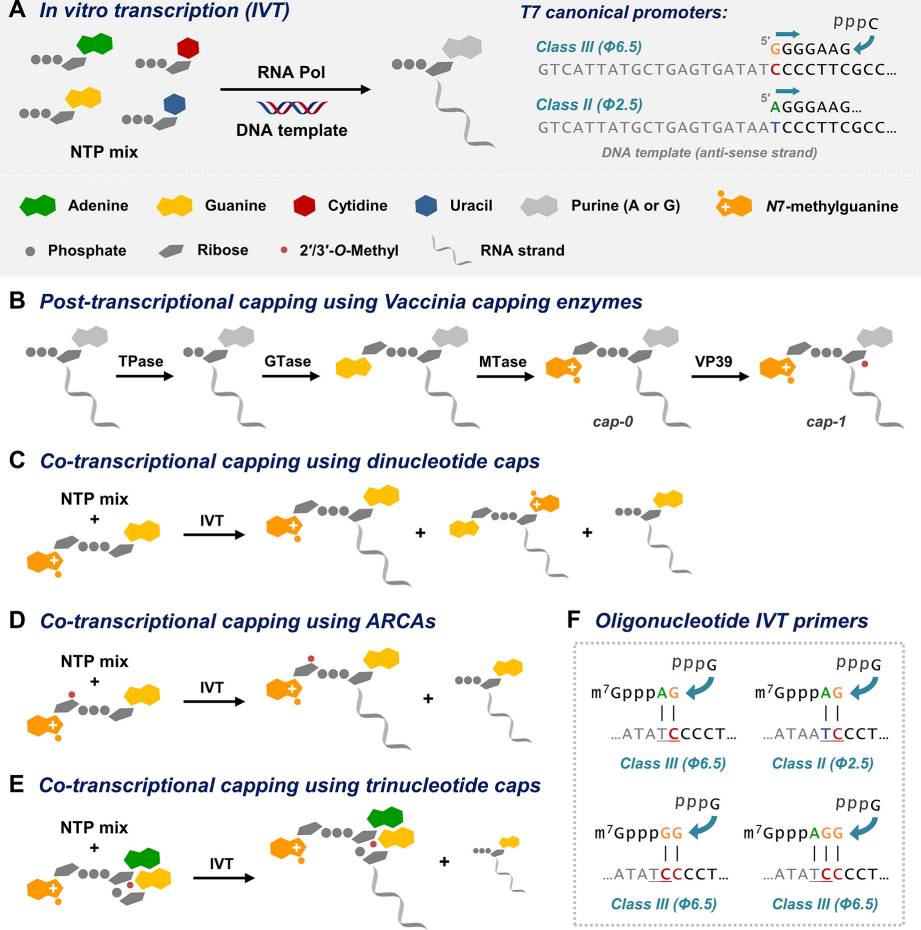 Fig. 2 Laboratory methods of mRNA synthesis and capping.2
Fig. 2 Laboratory methods of mRNA synthesis and capping.2
The post-transcriptional capping technique adds a 5′ cap structure to mRNA molecules subsequent to the completion of the IVT process. This technique synthesizes the mRNA strand without its cap structure and subsequently subjects it to enzyme treatment for cap addition. Guanylyltransferase and methyltransferase enzymes are used during the process to create both cap 0 and cap 1 structures. Researchers typically select Vaccinia virus capping enzyme (VCE) to synthesize cap 1 mRNA but choose Faustovirus capping enzyme (FCE) when they need a broader temperature range and higher enzyme activity. To obtain a cap 1 structure from cap 0 the mRNA Cap 2′-O-Methyltransferase (2′-O-MTase) enzyme must perform an additional reaction. This technique achieves excellent capping efficiency and exact control of cap structures which makes it ideal for situations requiring particular cap formations. The addition of enzymatic steps and purification procedures to the process extends the duration of the IVT procedure. The added steps involved in post-transcriptional capping do not diminish its importance as a method for generating high-quality, capped mRNA needed for both research and therapeutic applications.
IVT reaction processes that implement co-transcriptional capping directly incorporate cap analogs and bypass subsequent enzymatic treatment and purification stages. By utilizing this approach producers can achieve streamlined mRNA generation that results in high functional molecule output. CleanCap and ARCA are the two main technologies that scientists use for co-transcriptional capping.
CleanCap represents the newest evolution of cap analog technology. The technology enables cap-1 mRNA synthesis directly during transcription with greater than 95% efficiency and high production levels. It does not require a high cap: CleanCap maintains a more optimal balance during transcription without requiring a high cap:GTP ratio. CleanCap proves especially beneficial when used for therapeutic purposes.
ARCA creates cap-0 mRNA throughout the IVT reaction before needing mRNA cap 2′-O-methyltransferase to generate cap-1 mRNA. ARCA displays reduced capping efficiency that ranges between 50 to 80 percent when compared to CleanCap. The absence of licensing requirements in ARCA serves as a beneficial aspect for certain users.
The capping efficiency of IVT mRNA production remains a vital factor because it determines both the stability and translatability of the mRNA. Multiple techniques have been introduced to measure capping efficiency which present distinct benefits and shortcomings. A simple technique to measure capping efficiency involves attaching a fluorescent marker to the 5′ cap of the mRNA. The level of fluorescence emitted by the sample directly reflects how efficiently the IVT mRNA has been capped. Users can obtain a rapid evaluation of cap structures through this uncomplicated procedure. Ribozyme assays cleave IVT mRNA at a distinct position near the 5′ end to produce short 5′ cleavage products which may have either capped or uncapped ends. The purification of these products takes place through silica-based columns before visualization and quantification through denaturing polyacrylamide gel electrophoresis (PAGE) or liquid chromatography combined with mass spectrometry (LC–MS). The approach permits evaluation of capping efficiency in various synthetic mRNAs characterized by distinct cap structures with differences in 5′ untranslated regions, nucleoside modifications and variable lengths. The LC-MS technique serves to separate and determine the amounts of both capped and uncapped mRNA species. The method delivers precise and dependable results about capping efficiency and detects various byproducts created during the process in the sample mixture. The intact quantification workflow within SCIEX OS software delivers efficient data reduction capabilities that facilitate easier determination of capping efficiency.
Establishing quality testing protocols for capping procedures is vital to maintain the integrity and functionality of mRNA-based therapeutics. IVT templates are produced from linearized plasmids that carry the coding sequences between 5′ and 3′ UTRs with poly(A) tails. The linearization process ensures appropriate cap structure formation during mRNA transcription. The synthesis of IVT mRNA occurs through transcription kits while specific cap structures (cap 0 or cap 1) are formed by adding cap analogs such as ARCA-G or CleanCap. Vaccinia virus capping enzymes facilitate enzymatic capping to obtain either cap 0 or cap 1 mRNA structures. Ribozymes perform targeted cleavage on IVT mRNA at designated positions close to the 5′ end to produce short 5′ cleavage products. Silica-based columns are used to purify these products by eliminating long RNA fragments and ribozymes. Researchers visualize purified 5′ cleavage products by conducting 21% PAGE with 8 M urea or utilizing LC–MS analysis. Quantification of the bands representing capped and uncapped products enables the calculation of capping efficiency which depends on the relative intensities of these bands.
At BOC Sciences, we offer a comprehensive suite of mRNA capping services tailored to enhance transcript stability, translational efficiency, and immunogenicity control for your specific application. From conventional enzymatic methods to state-of-the-art co-transcriptional technologies, our custom mRNA capping solutions are designed to meet both research and clinical-grade needs.
We support both enzymatic and co-transcriptional strategies for the generation of:
We utilize high-purity cap analogs and enzymes (e.g., 2'-O-methyltransferase) to ensure efficient cap conversion and uniformity across your transcript batches.
For seamless and scalable production, we offer co-transcriptional capping using advanced cap analogs such as:
Our co-transcriptional workflows streamline production while delivering high capping efficiency and minimal byproduct formation.
We help you select and optimize cap structures based on:
We also offer flexibility in transcript lengths, incorporation of modified nucleotides, and inclusion of UTRs for optimal expression profiles.
All capped mRNA products undergo:
Whether you're optimizing a vaccine candidate, scaling a gene therapy construct, or developing next-gen mRNA tools, our custom capping solutions deliver performance, consistency, and regulatory readiness.
Partner with us to achieve high-quality, translation-ready mRNA with the capping strategy that best suits your needs.
We offer both enzymatic post-transcriptional capping and co-transcriptional capping using advanced analogs like CleanCap and ARCA to meet different research requirements and efficiency targets.
We employ multiple analytical methods including LC-MS, ribozyme assays, and electrophoresis to precisely quantify capping efficiency and ensure batch-to-batch consistency.
Selection depends on research application, target cell type, required translation efficiency, and immunogenicity considerations, with customized solutions for each project's specific needs.
Yes, we provide custom cap analog integration and optimization services to develop specialized cap structures tailored to unique experimental requirements and expression profiles.
Our comprehensive QC includes capping efficiency analysis, mRNA integrity assessment, dsRNA quantification, and functional validation to guarantee research-ready capped mRNA.


References