The outbreak of the COVID-19 disease caused by the RNA virus SARS-CoV-2 has become a large-scale epidemic. Surface glycoproteins and nucleocapsid phosphoproteins are two important proteins of the virus. They promote the virus's entry into host cells and genome replication.
Small interference RNA (siRNA) is a promising tool for RNA interference (RNAi) pathway. It inhibits viral gene expression by hybridizing and neutralizing target complementary degradation mRNA to control human virus infection. In order to slow the COVID-19 pandemic and restore affected individuals, the development of siRNA treatments may be a promising alternative to traditional vaccine design.
Recently, scientists from Dhaka University in Bangladesh, the University of New South Wales in Australia, and BRAC University in Bangladesh used RNA interference technology to develop siRNA molecules targeted specific target genes, namely nucleocapsid phosphoprotein gene and surface glycoprotein gene. The related result was published on April 12 in bioRxiv.
The researchers collected conserved sequences from 139 SARS-CoV-2 strains around the world and used computational methods to design and construct 78 siRNAs capable of inactivating nucleocapsid phosphoprotein and surface glycoprotein genes. And then, researchers used siRNAPred to check the effectiveness of the predicted siRNA. Finally, through a rigorous filtering process, 8 siRNA molecules were screened with exerts the best action.
The researchers said that during siRNA-mediated therapy, these predicted siRNAs should effectively silence the SARS-CoV-2 gene during siRNA-mediated therapy, thereby helping to resist SARS-CoV-2.
Although This study employed extensive computational methods to assess the quality of the designed siRNA sequences, which needs to be further explored in an in vitro laboratory environment.
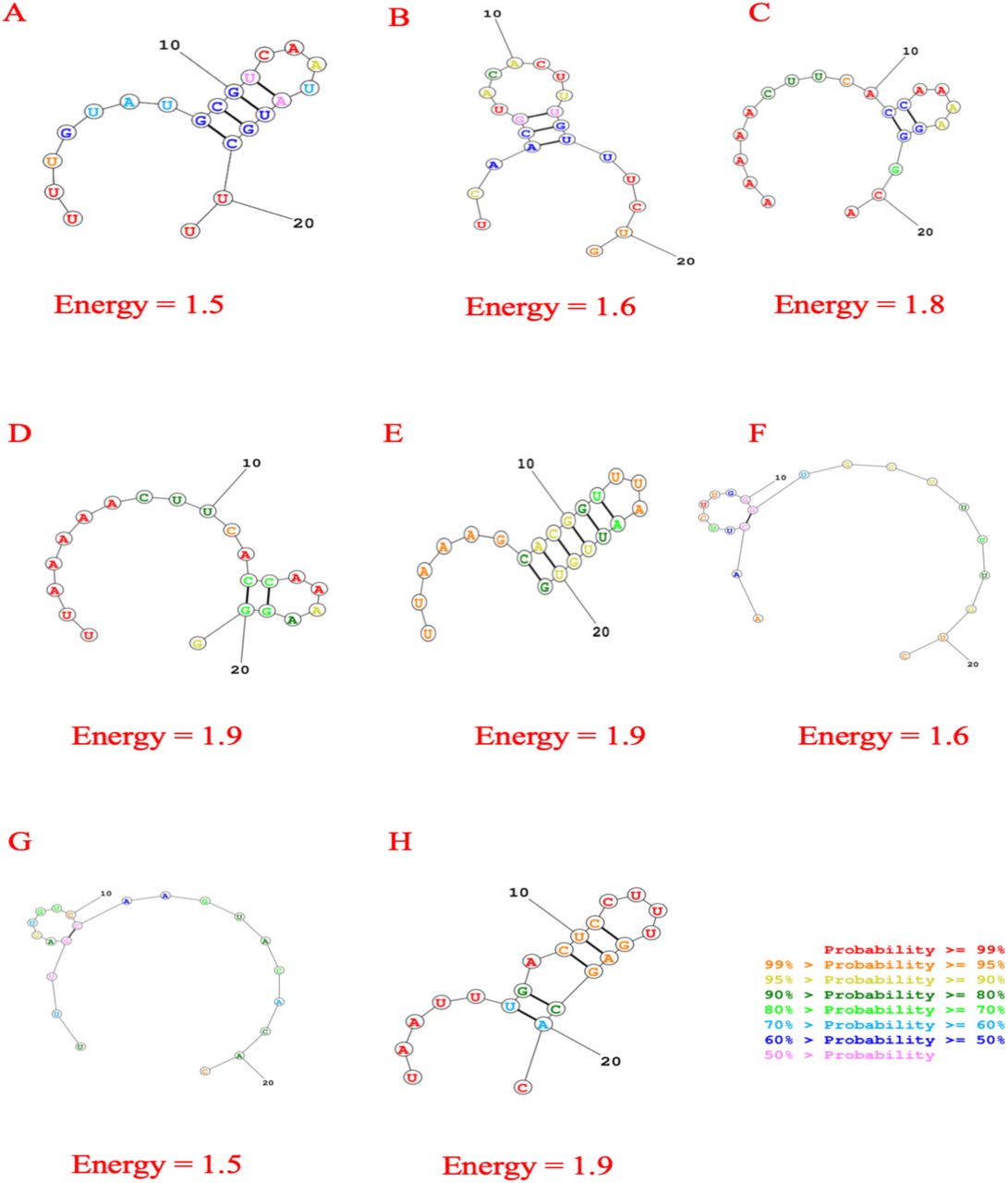 Figure 1: Secondary structures of best eight predicted siRNA with possible folding and minimum free energy for consensus sequence.
Figure 1: Secondary structures of best eight predicted siRNA with possible folding and minimum free energy for consensus sequence.
siRNA can specifically bind to complementary viral mRNA, such as nucleocapsid or surface glycoprotein genes, reducing their expression and providing a platform for viral gene functional studies.
Tools like siRNAPred and sequence conservation analysis allow researchers to predict potency, stability, and off-target effects, streamlining the selection of high-quality siRNA candidates.
By analyzing multiple viral genome sequences globally, conserved regions are identified to design siRNAs that maintain effectiveness despite viral mutations.
Validation involves confirming target mRNA knockdown, assessing sequence specificity, and optimizing delivery methods to maximize uptake and gene silencing in cell models.
Custom services provide sequence design, synthesis, quality control, and delivery optimization, reducing experimental time and supporting rapid exploration of viral gene function.
Yes, RNAi platforms can be adapted to silence genes of diverse RNA viruses, making it a versatile tool for studying viral replication, protein function, and host-virus interactions.
Ref: