5' capping is a type of post-transcriptional modification that controls the fate of mRNA and almost exclusively occurs co-transcriptionally in eukaryotic cells. It refers to the addition of a cap or modified nucleotide to the 5' end of an mRNA transcript. This modification is a key factor in the regulation of mRNA stability and translation. The addition of a cap at the 5' end of an mRNA transcript provides a protective feature against exonucleolytic attack. It also acts as a signal for translation initiation as it recruits the eukaryotic translation initiation factor 4E (eIF4E).
The 5' cap is a chemical modification to the 5' end of the mRNA, in which N7-methylguanosine is connected through a 5'-5' triphosphate bridge to the nascent transcript. The cap can be further methylated at the 2'-hydroxyl positions on the first (Cap 1) and second nucleotide (Cap 2) of the transcript. This additional methylation allows for innate immune evasion and an increase in translation efficiency. The cap is naturally associated with RNA polymerase II and is added co-transcriptionally by a capping enzyme complex that methylates guanine on N7 and ribose on 2'O to produce a structure that is identified as self by the translational and mRNA surveillance machinery of the cell. Synthetic mRNAs require a similar structure to prevent degradation and the induction of interferon. The core cap (Cap 0) contains the essential features of the cap structure—protection from 5' exonucleases and the binding of eIF4E—whereas Cap 1 and Cap 2 provide a stepwise addition of innate immune evasion, including resistance to IFIT1 and DXO, respectively. This hierarchy of structures provides a knob to tune the balance between increased stability and reduced immunogenicity on one hand and increased manufacturing complexity and expense on the other. This consideration impacts nearly all other decisions in mRNA formulation, including the selection of a cap analog, as well as the optimisation of IVT conditions to ensure high capping efficiency and transcript integrity.
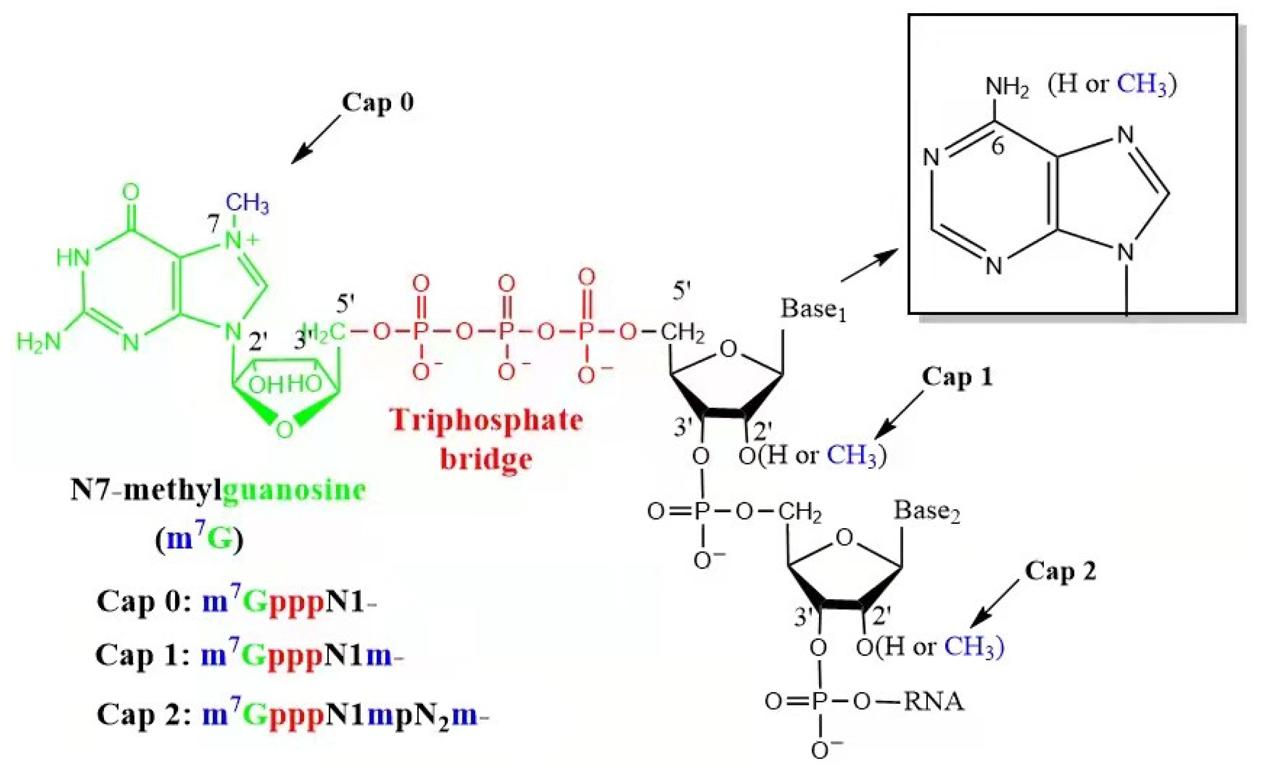 The types of an mRNA cap. IVT mRNA contains three types of caps, i.e., Cap 0, Cap 1, and Cap 2.1,5
The types of an mRNA cap. IVT mRNA contains three types of caps, i.e., Cap 0, Cap 1, and Cap 2.1,5
The 5' cap structure of mRNA is an unusual nucleotide arrangement in eukaryotes with an inverted 5'–5' triphosphate linkage. It is the first nucleotide at the 5' terminus of the transcript, but this nucleotide is linked in reverse orientation (chemically: 5' to 5' instead of 5' to 3') to the first transcribed nucleotide residue, which thereby becomes a substrate and part of the cap structure. This connection of the first residue by the triphosphate bridge to the terminal guanosine nucleotide effectively masks the 5' end from 5'-to-3' exonucleases, which otherwise would quickly degrade the molecule. The simplest form of cap structure, cap 0, is the m7G residue itself in a 5' to 5' triphosphate linkage to the first nucleoside of the RNA transcript. In most eukaryotes this structure alone is sufficient to provide resistance to degradation and translational competence. In higher eukaryotes this is further methylated in a 2'-O-methylation on the riboses of the first and second nucleotide of the RNA transcript, creating structures that are referred to as cap 1 and cap 2, respectively. The latter two structures are recognized as self by the innate immune system, which inhibits inappropriate activation of anti-viral responses and also increases binding affinity for eukaryotic translation initiation factor 4E. The phosphate bridge connecting the first and the cap nucleotide can have more than three phosphate units, for example a tetraphosphate bridge which is reported to have higher stability and enhanced protein binding affinity. The three-dimensional structure of the cap is predicted to form specific base stacking between the 7-methylguanosine cap and the first two transcribed residues.
Table 1 Structural Hierarchy of mRNA Caps.
| Cap Type | Methylated Components | Structural Formula | Biological Distribution | Key Functional Attributes |
| Cap 0 | N7-methylguanosine only | m7GpppN | Yeast, plants, invertebrates | Basic stability and translation |
| Cap 1 | N7-methylguanosine + first nucleotide 2'-O-methyl | m7GpppNmN | Mammalian mRNA | Immune evasion, enhanced translation |
| Cap 2 | N7-methylguanosine + first two nucleotides 2'-O-methyl | m7GpppNmNm | Select mammalian transcripts | Maximum self-RNA identity |
| TMG cap | 2,2,7-trimethylguanosine | m2,2,7GpppN | snRNAs, some viral mRNAs | Nuclear localization, specialized function |
The consequences of capping and uncapping are dramatic: in most organisms, the capped transcript is protected from nuclease attack, translated efficiently, and invisible to the immune system, whereas the uncapped message is promptly degraded, untranslatable, and the strongest known inducer of the interferon response. Lacking both the N7-methylguanosine cap as well as 5'-5' triphosphate linkage, the uncapped transcript features a free 5'-hydroxyl end immediately recognized by 5'–3' exonuclease XRN1 as well as other 5'–3' decay machineries. It has half-life of minutes, as opposed to hours for the capped version. In addition, the unmasked 5' terminus strongly activates interferon pathway: RIG-I directly binds 5'-triphosphate and triggers MAVS and downstream IRF3/7 activation that culminates in production of type I interferon while IFIT1 sequesters eIF4E from its target by competing for cap binding and therefore completely halts translation initiation. Capped mRNAs, in contrast, recruit eIF4E through the N7-methylguanosine cap, which assembles eIF4F complex and licenses 43S ribosome scanning, thereby allowing translation initiation to commence at the correct AUG. In addition, the cap protects transcript from decapping enzymes such as DCP2 and DXO, whose decapping is required to initiate mRNA degradation, whereas the uncapped version is degraded immediately.
The 5' cap serves as an active defense against both constitutive and inducible mRNA decay, defining the temporal half-life of the transcript in cell and body. It does so in three ways: by preventing access of 5'-to-3' exonucleases; by recruiting cap-binding proteins which sterically prevent endonucleolytic cleavage; and by serving as the binding platform for poly(A)-binding protein, which circularises the transcript and provides synergistic stability to both ends of the mRNA. Loss of the cap (spontaneous hydrolysis or decapping) leaves the 5' end open to rapid decay. The newly-exposed 5' monophosphate becomes a substrate for XRN1-mediated exonucleolysis, and the poly(A) tail becomes deadenylated by CCR4-NOT and PAN2-PAN3. The stability of the cap is therefore a quality attribute of therapeutic mRNA. Features that increase resistance to hydrolysis and decapping, such as phosphorothioate bridges or 2'-O-methylation, can prolong the half-life of a transcript from hours to days and reduce the frequency of dosing required to achieve a therapeutic protein level. The cap can also modulate sensitivity to RNase L, an interferon-induced endoribonuclease which cleaves uncapped or incompletely capped transcripts in an antiviral response, highlighting the importance of the cap not only for stability, but also for safety.
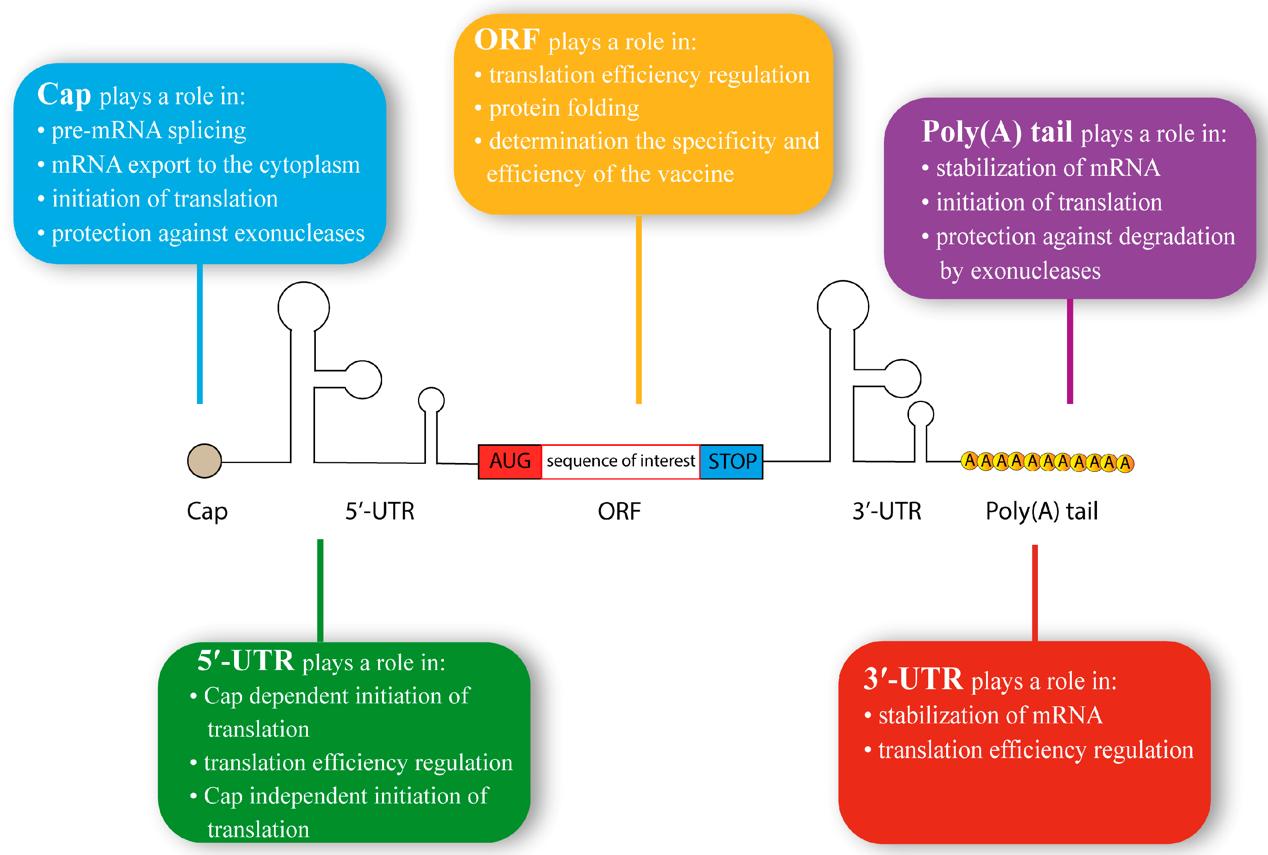 The typical structure of synthetic mRNA.2,5
The typical structure of synthetic mRNA.2,5
The cap structure constitutes a strong steric barrier against 5'–3' exonucleases. In the absence of the cap, 5'–3' exonucleases would otherwise start degradation of messenger RNA from the very end. The 5' cap has an inverted 5'–5' triphosphate linkage which forms a distinct chemical junction that 5'–3' exonucleases are unable to accommodate in their active site. Processive 5'–3' exonucleases like Xrn1 are not able to move across this chemical junction due to steric clashes by the methylated guanosine with the active site of the enzyme. This means that the 5' cap will prevent the 5'–3' exonuclease from even accessing the phosphodiester backbone of the transcript. The cap also has a different distribution of negative charge compared to the exposed 5' triphosphate or monophosphate of uncapped or decapped transcripts and is less electrostatically attracted to exonuclease binding domains as a result. The 5' cap structure also blocks the entry into processing bodies, degradation foci of non-protected RNA. 5'–3' exonucleolytic degradation is also blocked from an early stage at the nucleus by the 5' cap which blocks Rat1-mediated degradation of incompletely processed transcripts. This protection is in place throughout mRNA life, as the 5' cap is important in blocking degradation during mRNA nuclear export, cytoplasmic transport and during multiple rounds of translation.
The 5' cap serves as a molecular platform for the recruitment of cap-binding proteins. These proteins associate to form ribonucleoprotein complexes that are capable of actively influencing mRNA stability, and modulating the overall mRNA metabolic fate. In the nucleus, the CBP80/20 heterodimeric complex is directly recruited to the cap and is involved in the coupling of mRNA processing events. It also protects the transcript from degradation before processing is complete. In the cytoplasm, the nuclear cap-binding complex is replaced by the eIF4E. eIF4E binds the cap and recruits additional proteins to form the eIF4F complex, which protects mRNA during and after translation initiation. Binding of these proteins to the cap creates a steric barrier that also protects the entire 5' region. Cap-binding proteins also act as adaptors that tether capped transcripts to other mRNA stability factors including poly(A)-binding protein (PABP) to create pseudo-circularized structures that protect both the 5' and 3' ends simultaneously. In addition, some cap-binding proteins have an enzymatic function that senses the integrity of the cap and removes improperly modified caps, which are then targeted for degradation. The decapping enzymes, Dcp2, that are normally excluded from the cap structure, become recruited after deadenylation to promote regulated decay of the transcript.
A key feature of a correctly formed 5' cap structure is that it increases the half-life of a transcript. A transcript that would otherwise be short-lived is instead able to persist, potentially over the course of days. This persistence is due to a number of protective mechanisms acting in concert to prevent both targeted and stochastic degradation. Exonuclease resistance, for example, acts as a simple barrier to prevent ongoing enzymatic decay; in other words, capped transcripts will be far more stable in the nucleocytoplasmic milieu than uncapped ones. A complementary mechanism is protection by protein binding, whereby associated proteins shield the transcript even when stress occurs. Capped transcripts are inherently more stable because of their role in translation; actively translating mRNAs have ribosomes bound which sterically hinders access to endonucleases and decay machineries. Proteins bound to the cap structure may also protect transcripts from prematurely entering processing bodies, which are cytoplasmic granules that recruit uncapped transcripts for decay. Half-life regulation of a transcript can thus be affected by cap status, since a transcript can be actively destabilized by decapping if the protein product is no longer needed. For synthetic mRNA therapeutics, the duration of action is directly controlled by the cap, since a more stable cap structure will result in the protein being expressed for a longer period, potentially a matter of days rather than hours, and thus the dose may be reduced in frequency.
Table 2 Molecular Mechanisms Linking Cap Structure to mRNA Stability.
| Stability Mechanism | Molecular Basis | Functional Outcome | Influence on Half-Life |
| Physical barrier | Inverted 5'–5' triphosphate linkage blocks exonuclease access | Prevents 5'-to-3' degradation | Dramatic extension |
| Protein shielding | Recruitment of cap-binding proteins | Steric protection of 5' region | Moderate to substantial extension |
| Immune evasion | 2'-O-methylation prevents pattern recognition receptor binding | Avoids targeted destruction | Substantial extension in mammalian systems |
| Translation coupling | Efficient ribosome recruitment and scanning | Ribosomal protection during translation | Variable, depends on translation rate |
| Compartmental exclusion | Cap prevents entry into processing bodies | Avoids sequestration in decay centers | Moderate extension |
| Regulated decay | Controlled decapping after deadenylation | Timely transcript removal | Programmed reduction when needed |
The 5' cap structure serves as the master identity card that allows eukaryotic mRNA access into translation. By virtue of presenting a 7-methylguanosine moiety in an inverted 5'–5' triphosphate linkage, the cap precludes exonucleolytic decay and recruits the cap-binding complex eIF4F to nucleate the 43S pre-initiation complex assembly. In its absence, transcripts are translationally inert and rapidly degraded. The cap is therefore a checkpoint that converts the flow of genetic information into protein output. Stability is therefore a hardwired design constraint for synthetic mRNA therapeutics.
The cap is initially recognized by eIF4E, whose positively charged surface is docked onto the methylated guanine. This induces a conformational change that in turn exposes its binding site for eIF4G. The large scaffold protein, in turn, tethers eIF4A (an ATP-dependent helicase) and eIF3 (the 43S ribosomal anchor). The eIF4F complex thus serves as a molecular bridge that physically links the small ribosomal subunit to the mRNA. Since the cap is outside of the ribosomal footprint, this assembly automatically positions the 43S complex at the extreme 5' end, which in turn ensures that scanning will commence at the first AUG rather than at downstream, out-of-frame start codons. The entire recruitment cascade is therefore cap-dependent: any erosion or mis-orientation of the cap severs this bridge and abolishes translation initiation.
Once tethered, eIF4A then unwinds secondary structure in the 5'UTR, to enable the ribosome to scan downstream. The cap places a directionality constraint on this process: since initiation can only occur at the 5' end, the ribosome is forced to scan the entire 5'UTR, affording it many opportunities to discard near-cognate start codons. Only when the anticodon of Met-tRNAi base-pairs with the genuine AUG is eIF2-GTP hydrolyzed, the initiation factors released, and the 60S subunit recruited to form the 80S elongation-competent ribosome. The cap, then, does not just recruit the ribosome, but also properly positions it, so that translation commences at the correct start site and full-length protein is produced.
The cap also has effects post-initiation. By binding eIF4F, the cap also indirectly circularizes the mRNA through association with the poly(A)-binding protein. This provides increased ribosome processivity, and decreased drop-off during elongation. Forward-oriented caps (ARCA, Cap 1) have higher affinity for eIF4E and result in more initiations per transcript, as well as a longer half-life in function. Uncapped or caps in reverse orientation are quickly decapped and degraded, and even highly abundant RNA will express only an insignificant amount of protein. The result is that optimization of the cap (orientation, methylation status, and phosphate backbone) has a direct impact on dose-sparing, sustained expression, and lower cost of goods for therapeutics or vaccines.
Uncapped or partially capped synthetic mRNA is considered a quality defect. A cap that is missing, inverted, or not fully processed affects the functional performance and safety of a given transcript. The terminal cap structure is critical to protect mRNA from degradation and to initiate translation. A cap-defective mRNA, therefore, can be rapidly degraded and often has suboptimal translation efficiency, and may activate the innate immune system, with both aspects negatively affecting the performance and safety of an mRNA-based product. The degree of this problem will likely be correlated to the level of the quality defect. For instance, a fully uncapped mRNA will have a more detrimental effect than a partially capped mRNA which may still be active, yet has an increased potential to activate the innate immune system.
Decreased translation efficiency is the most immediate and measurable impact of uncapped or poorly capped transcripts. Translation is decreased by a variety of mechanisms. Firstly, uncapped or poorly capped mRNAs fail to recruit eIF4E, which specifically binds the 7-methylguanosine cap and initiates translation complex formation. As such, without cap recognition, ribosomes are not able to bind to mRNA and translation is nearly abolished. If the transcript is only partially capped, it may be bound by proteins such as IFIT1, which recognizes unmodified 5' ends of transcripts, thus sterically inhibiting translation initiation. Furthermore, mixed orientation caps (caps added to the transcript with 50% of them in the wrong orientation) are still chemically present on the mRNA; however, the translation machinery is unable to recognize the cap, thus these mRNAs are also translationally deficient. The impact of these mechanisms is that uncapped or partially capped transcripts experience a massive drop in protein expression (barely detectable levels), that is persistent throughout the experiment (the protective effect of translation on mRNA stability does not occur). This reduction in translation efficiency is also cell-type dependent, with cells such as professional antigen presenting cells being very sensitive to uncapped transcripts.
The second major effect of incorrect capping is enhanced mRNA degradation, which multiplies the translation defect by decreasing the half-life of the resulting synthetic transcripts. The presence of a cap is the first line of defense against the activity of 5'-to-3' exonucleases, especially of the main mRNA decay complex Xrn1. The decay occurs very quickly (half-life of minutes rather than hours) and thus any therapeutic window for protein production is abolished by this phenomenon. Uncapped transcripts are recognized by decapping enzymes Dcp1/Dcp2, which cleave the remaining cap or the 5'-linked nucleotide and leave the transcript accessible to Xrn1 exonuclease. Furthermore, incomplete capping results in accumulation of 5'-triphosphate groups that are recognized as degradation signals by the quality control systems in the cell, which identify such molecules as foreign, aberrant or viral. Moreover, another cause for the lack of stability is the absence of translational protection, when ribosome passage along coding sequence sterically shields the mRNA from exonuclease degradation. The rapid decay of uncapped or improperly capped mRNA is a cascading effect where not only is the translation less efficient, but the template itself is also rapidly degraded, allowing no accumulation of the protein product over time. The decay of these improperly capped RNAs is also sequence-dependent, with specific sequences of mRNAs being particularly sensitive to decay due to secondary structures that may be formed and may facilitate exonuclease access. Furthermore, the degradation rates of uncapped transcripts can also be different depending on the cellular state. For example, activated immune cells degrade such transcripts particularly quickly as a defense mechanism against viral RNAs.
Induction of an innate immune response is the most severe outcome of uncapping, as it can initiate an inflammatory cascade that both impairs the therapeutic effect and can cause safety issues. The innate immune system has evolved to sense uncapped RNA as a sign of viral infection, with several pattern recognition receptors that are tuned to recognize aberrant 5' ends. The primary sensor of uncapped RNA is the RIG-I receptor, which directly recognizes 5'-triphosphate groups that are exposed in the absence of a cap. RIG-I activation leads to a signaling cascade that results in strong production of type I interferon and pro-inflammatory cytokines, establishing an antiviral state that globally shuts down protein synthesis through various mechanisms. MDA5 is a second sensor of uncapped RNA that recognizes longer stretches of uncapped RNA, and which can also contribute to the inflammatory response. The inflammatory response to uncapped RNA is of particular concern for therapeutic applications, as the interferon response induces a number of proteins that target uncapped RNA for degradation while also globally shutting off protein synthesis. This includes, for example, IFIT1, which binds uncapped mRNA, and PKR, which phosphorylates eIF2α to globally shut off translation initiation. The magnitude of this inflammatory response can be significant, with reports that even low levels of contamination with uncapped mRNA can induce robust immune activation that completely prevents therapeutic protein expression. The response to uncapped RNA also has some cell-type specificity, with professional antigen-presenting cells in particular being more sensitive to uncapped RNA, likely reflecting their role in virus sensing. This can also have safety implications, as the inflammatory response can lead to local and systemic off-target effects.
Efficient 5' capping is a critical determinant of mRNA stability and translation efficiency, and it begins with the quality and design of the cap analog. Our cap analogs are specifically developed to support high-efficiency 5' capping during in vitro transcription (IVT), enabling reliable incorporation, robust protection against degradation, and effective interaction with the cellular translation machinery. Each product is optimized for compatibility with standard IVT systems and reproducible performance across different mRNA workflows.
Our cap analogs are engineered to deliver high capping efficiency during co-transcriptional IVT, ensuring a high proportion of properly capped mRNA molecules. By promoting correct cap incorporation at the 5' end, these reagents help improve resistance to exonuclease-mediated degradation and support efficient recruitment of translation initiation factors. This optimized capping performance contributes directly to improved protein expression and experimental reproducibility in mRNA-based systems.
Consistent mRNA performance depends on reliable raw material quality. Our cap analogs are manufactured under tightly controlled conditions to achieve high chemical purity, low impurity profiles, and strong batch-to-batch consistency. Rigorous analytical characterization ensures that each batch performs predictably in IVT reactions, supporting stable mRNA yield, integrity, and translation efficiency across research and advanced mRNA production workflows.
Proper 5' capping is essential for protecting mRNA from degradation and enabling efficient translation, and selecting the right cap analog can significantly improve both stability and expression outcomes. Whether you are optimizing an IVT workflow or troubleshooting performance limitations, our cap analog solutions are designed to support reliable and efficient 5' capping—contact us today to discuss your mRNA project and request technical guidance or a quotation.
References
Uncapped mRNA is rapidly degraded and shows very low translation efficiency.
The cap blocks exonuclease access and stabilizes mRNA structure.
Yes, the 5' cap is essential for recruiting translation initiation factors.
Proper capping significantly increases protein expression output.
Yes, incomplete capping leads to unstable mRNA and inconsistent translation.

