Cap analogs are artificial nucleotide units which mimic the 5' end of a eukaryotic messenger RNA. They are very useful in modern molecular biology, especially for in vitro transcription systems, and they can be added co-transcriptionally to the RNA as it is being transcribed. The 7-methylguanosine unit is connected to the first nucleotide by a 5',5'-triphosphate linkage, which mimics the natural cap structure which is important in post-transcriptional modifications, nuclear export, and translation.
Synthetic cap analogs (SCAs) have been a major area of research to overcome this need for the appropriate 5' modification. The first capping reaction must be performed on the transcript to produce a functionally active mRNA, as the cap is essential for mRNA half-life, transport, and translation via recognition of the cap-binding complex (CBC), and is primarily recognized by eukaryotic translation initiation factor 4E (eIF4E). Current in vitro transcription reactions (IVTs) made with bacteriophage polymerases can be a mix of correctly and incorrectly oriented cap structures, which can reduce the activity of the RNA. This is because the polymerase can use either the 5'-OH or 3'-OH of the dinucleotide cap donor, leaving transcripts with the methylated guanosine in the non-functional position. The most recent work has moved toward the use of anti-reverse cap analogs (ARCAs) with steric hindrance to produce only 5' to 3' transcripts, resulting in more uniform capping and improved translation activity. These modifications have contributed to the development of new, more potent, longer-acting, and less immunogenic mRNA therapeutics and vaccines.
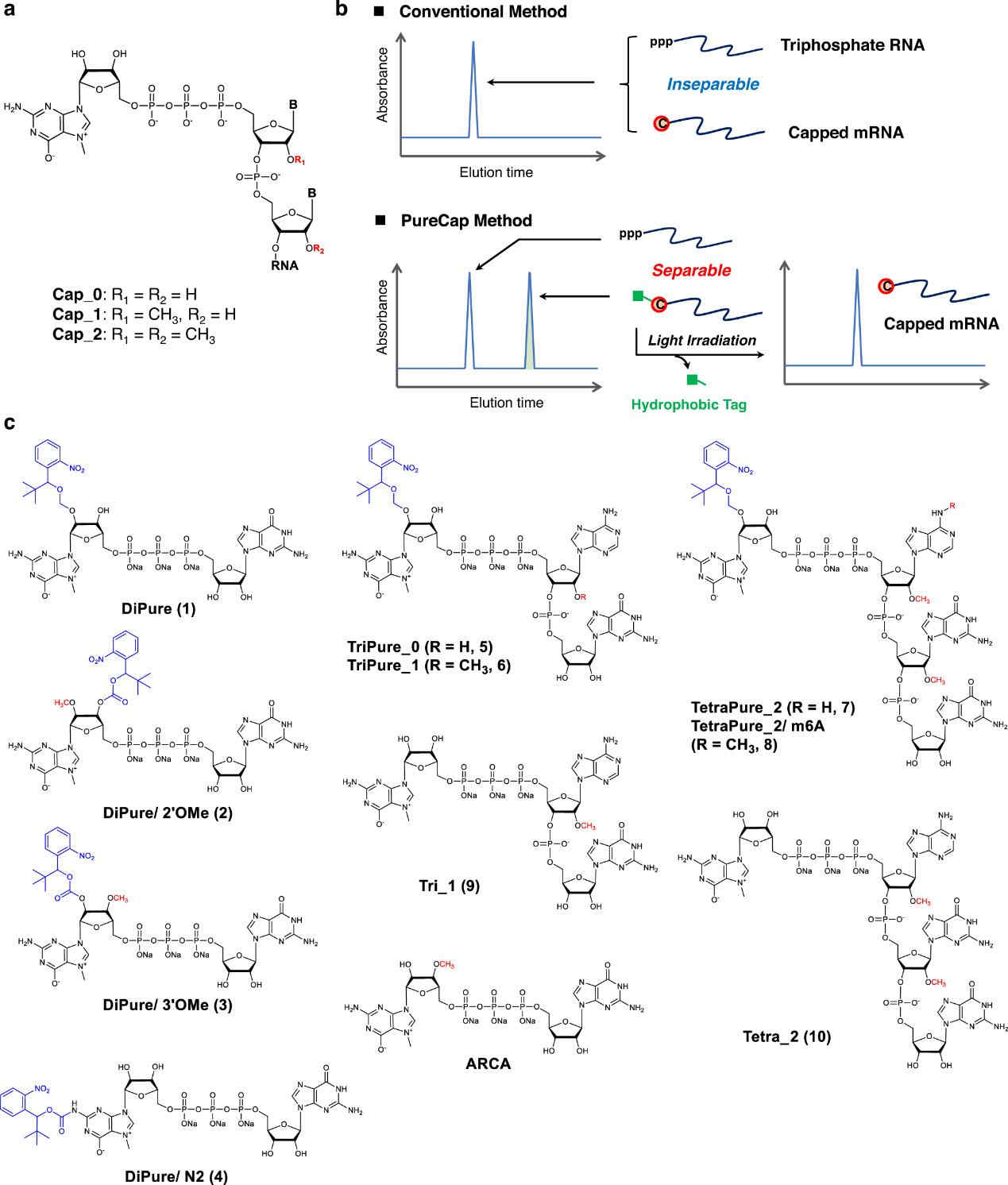 Hydrophobic cap analogs for the purification of capped mRNA using RP-HPLC (the PureCap method).1,5
Hydrophobic cap analogs for the purification of capped mRNA using RP-HPLC (the PureCap method).1,5
A cap analog, also termed an artificial cap or synthetic cap, is a dinucleotide or oligonucleotide molecule that is used in place of the 5' cap in vitro. It is structurally similar to the natural 5' cap. Cap analogs are most often used during the in vitro transcription (IVT) process. Cap analogs can be described as a 7-methylguanosine residue connected to the first initiation nucleoside by an inverted 5'–5' oligophosphate linkage. They are represented as m7GpppN, where N represents the initiating nucleoside that is chemically linked to the 7-methylguanosine (m7G) via the 5' phosphate. Cap analogs can be used by RNA polymerases as a substrate in IVT reactions and are added to the 5' end of the IVT product. Cap analogs typically have chemical modifications (such as a modification of a ribose hydroxyl group or a modification to the phosphate backbone) that allow them to only be added to the 5' end of RNA in the forward direction. This process allows mRNA to be translationally activated and protected without the need for enzymatic capping after IVT.
The natural cap structure is a 7-methylguanosine residue added co-transcriptionally through a complex interplay of nuclear enzymes, and as such has 100% incorporation and natural patterns of methylation (cap 0, cap 1, and cap 2). The natural cap structure was optimized to finely regulate translation initiation and degradation of mRNAs. Artificial cap analogs have the same core structure, but their synthesis is often optimized for cost and different desired properties. The first artificial cap analogs could be added in either orientation, but methylation of the hydroxyl group at the 5' end directs addition in the only correct 5'–5' orientation. Synthetic cap analogs are often derived from tetraphosphate linkages (instead of the natural triphosphate linkage), which have improved stability and affinity for eIF4E. The natural capping mechanism ensures that all transcripts are capped, but artificial capping competes with GTP in stoichiometric ratios, and as such often only partially caps transcripts. Newer cap analogs can have much greater incorporation efficiencies, as well as properties not easily achievable with natural capping, such as resistance to decapping or direct synthesis of cap 1.
Table 1 Comparative Features of Natural and Synthetic mRNA Caps.
| Feature | Natural mRNA Cap | Synthetic Cap Analogs |
| Installation mechanism | Enzyme-catalyzed co-transcriptional modification | Chemical incorporation during IVT |
| Fidelity | Complete modification of all transcripts | Efficiency varies by analog design |
| Orientation control | Inherent enzymatic directionality | Requires chemical modifications to prevent reverse incorporation |
| Structural variants | Cap 0, cap 1, cap 2 (regulated) | Can be engineered for specific structures (cap 0, cap 1, extended phosphate) |
| Scalability | Limited to cellular production rates | Suitable for large-scale manufacturing |
| Customizability | Fixed biological pathway | Tunable stability, translation efficiency, immunogenicity |
One of the main reasons cap analogs are used in in vitro transcription workflows is that it allows for a single step in vitro cap installation during transcription (called co-transcriptional capping). This is a more streamlined manufacturing approach compared to enzymatic multi-step capping workflows as it does not require a separate capping step. This results in shorter production time, less material loss in the purification steps, and ultimately lower costs, with potential for large scale manufacturing advantages. The simplified workflow is essential for large scale manufacturing of mRNA when used for therapy or prophylaxis, where efficient process development and minimal batch-to-batch variability are highly important. Moreover, synthetic cap analogs can have a range of tunable properties, not easily achieved by enzymatic capping approaches: a wide range of chemical modifications with different properties that influence translation efficiency, stability, and innate immune recognition can be utilized, and the final cap structure can be designed to optimize for specific features. For example, cap analogs that directly form a cap 1 structure without a further methylation step can be used to simplify workflows and reduce double-stranded RNA impurities that can activate innate immune receptors. Resistance against decapping enzymes can also be engineered, to extend mRNA half-life in the cell, to maximize the protein output per mRNA molecule. While early cap analogs had lower incorporation efficiencies, modern cap analogs can reach high capping efficiencies even at low concentrations.
The cap analog is the molecular primer that determines the fate of IVT mRNA: it establishes the 5'-terminal architecture at the very beginning of chain elongation, which effectively couples cap installation to polymerase activity, thereby obviating the need for a post-synthetic modification step. The identity of the analog also has profound impact on IVT transcription yield, capping efficiency, orientation fidelity and downstream immunogenicity, and is thus a key determinant of both process robustness and clinical performance. In contrast to natural transcription, where capping enzymes modify the nascent chain after initiation, the cap analog is required to act as both a substrate for the polymerase active site as well as a ligand for downstream translation factors. This dual requirement effectively limits chemical design and necessitates careful optimization of its concentration ratios with respect to the NTPs.
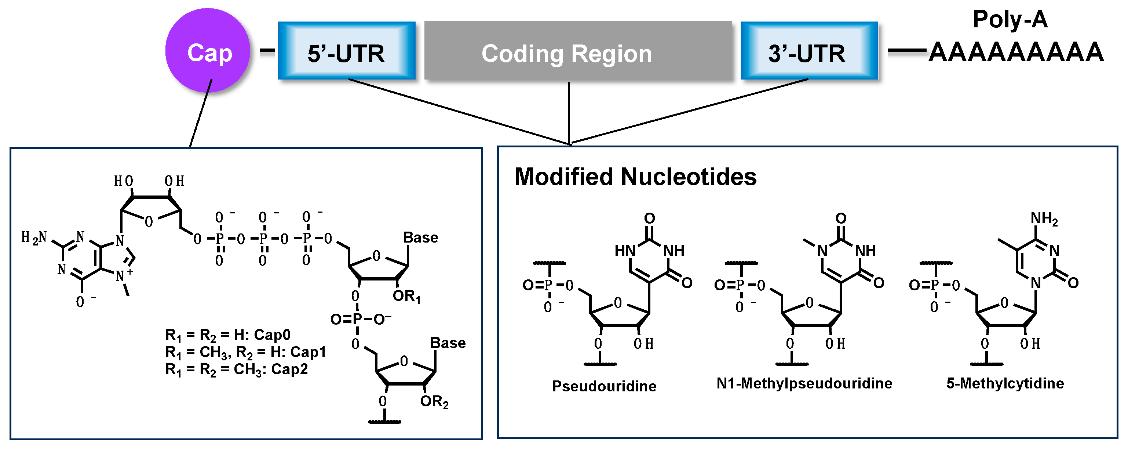 Structure and established chemical modifications of mRNA.2,5
Structure and established chemical modifications of mRNA.2,5
Cap analogs are added to newly transcribed RNA by competing with GTP at the time of initiation of transcription. The T7, SP6 and T3 RNA polymerases, which are isolated from bacteriophages, bind to the promoter template and add a cap dinucleotide (or a regular nucleotide) to the 5' end of the transcript. First-generation cap analogs like m7GpppG, have directionality issues, as the RNA polymerase can extend the chain from either 5' or 3' hydroxyl group of the dinucleotide. As a result, the capped RNA is a mixture of correct and reverse orientation capped RNAs. Reverse transcriptase inhibitors (anti-reverse cap analogs) are chemically modified such that the chain cannot be extended from the methylated guanosine. As a result, the RNA transcripts are all in the right orientation and nearly 100% of the capped transcripts are able to bind to eIF4E. Co-transcriptional capping results in synthesis and modification of RNA in one reaction step, which is less error prone and has fewer contamination issues. It also allows the user to directly control the ratio of different capped structures by adjusting the concentration of the cap analog in the reaction mix.
The cap analog competes with the normal initiating nucleotide and it must be recognized by the RNA polymerase. This binding of cap analogs is in the active site of the RNA polymerase. The 7-methylguanosine unit of the cap is further upstream of the dinucleotide and the substrate tolerance of the enzyme allows this recognition to occur. If one were to change the structure of the cap analog it would change the efficiency with which it was incorporated into the initiation complex, since it would change how well it was bound to the active site. The anti-reverse cap analog, in which 2'-O-methylations are present on the ribose sugar prevents addition of a nucleotide at the modified guanosine, and thus ensures the reaction only proceeds in the proper direction. The binding of the analogs also depend on their structure; phosphorothioate modifications can change the kinetics of the reaction. The reaction can also be controlled by altering the concentration of the analog and the competing nucleotide, as well as the conditions of the reaction (magnesium ion concentration, temperature, etc.), which will alter the overall capping efficiency in the final product.
Synthetic cap analogs are also used in the post-transcriptional enzymatic capping process that occurs separately from the RNA polymerization reaction. This method generally uses a capping enzyme to convert the 5' triphosphate to a diphosphate and then install a 7-methylguanosine cap with S-adenosylmethionine. This multi-step enzymatic reaction necessitates additional purification between steps, which lengthens production time and cost, as well as increasing the potential for batch-to-batch variation. Although enzymatically capped products have the correct orientation, such capping requires more optimization and quality control, as multiple enzymes are needed to install the cap and, if desired, a Cap-1 structure with 2'-O-methylation. Cap analogs also allow for the product to be made in a single reaction vessel, decreasing potential contamination and reducing process complexity. Anti-reverse cap analogs with modern designs can also achieve near-enzymatic orientation purity but with some structural changes that can improve their stability or translation, such as phosphorothioate linkage or longer phosphates. Co-transcriptional capping is more scalable and rapid, while post-transcriptional capping is less common and is sometimes required for specialized applications where control of the number and position of cap methylation is needed.
Cap analogs are divided on the basis of their structure into types and subtypes depending on their functional and methylation attributes. The most important division is based on the extent of the 2'-O-methylation of the 1st and 2nd transcribed nucleotide. The three most common of these cap types are cap 0, cap 1, and cap 2. Cap analogs are also differentiated based on the addition of an orientation control mechanism which allows only one-way growth of the transcript, called anti-reverse cap analogs. Other distinctions can also be made based on the length of the phosphate bridge between the nucleotides and the substitution of certain groups within the analog structure, which can increase stability and the affinity to certain proteins. This wide range of different structural cap analogs allows for the appropriate reagent to be chosen for a specific application based on a tradeoff of translational potency, immunogenicity, and ease of production. As the biology of the cap became more understood more complex structures have been synthesized, with dinucleotide cap structures evolving into modified cap structures.
Cap 0 analogs are the simplest cap structures, with the N7-methylguanosine linked to the first transcript nucleotide with a 5'-5' triphosphate bridge (m7GpppN). This simple structure affords basic protection from 5'-exonucleases, but is not protected from cytosolic pattern-recognition receptors like IFIT1 which bind to the unmethylated 2'-hydroxyl of the first nucleotide and thereby inhibit translation initiation, while also activating interferon responses. Thus, mRNA capped solely with Cap 0 is poorly translated in mammalian cells and quickly degraded in the presence of serum RNases. The main disadvantage of dinucleotide Cap 0 reagents is orientation indeterminacy: in in-vitro transcription, the cap will be added in either the forward or reverse orientation, and the reverse orientation is non-functional with respect to translation, lowering protein yield by 30-50%. As such, Cap 0 analogs are most useful for basic studies of innate immunity, cancer immunotherapy applications in which interferon induction is a goal, and in high-throughput screening applications in which price trumps performance. They are also the easiest to manufacture, being produced in multimolar quantities at low cost with simple phosphorylation chemistry, and as such are very cost effective for preclinical proof-of-concept studies.
Cap 1 and Cap 2 analogs have 2'-O-methylation on the first and second transcribed nucleotides, respectively, and are more similar in structure to naturally occurring mammalian mRNA. They are less immunogenic and do not activate the pattern recognition receptors RIG-I or MDA5 (RNA sensors that detect self versus non-self RNA based on 2'-O-methylation patterns), while the additional methyl groups have a higher affinity for eIF4E and are more resistant to some decapping enzymes, leading to longer half-lives and higher protein expression. Cap 1 analogs are now considered standard for therapeutic mRNAs, as they are more similar to naturally occurring mRNA structures and balance translation efficiency with reduced immunogenicity. Cap 2 structures can also be made, but their increased stability may not be worth the more difficult synthesis depending on the application. In the past, Cap 1 required stepwise enzymatic methylation following transcription. Cap 1 and Cap 2 structures are now easily made using trinucleotide and tetranucleotide analogs that directly incorporate pre-methylated nucleotides during synthesis, leading to improved batch-to-batch consistency. This has been key for the use of mRNA in vaccine development as well as for protein replacement therapies, where a high expression with minimal inflammation is required.
Table 2 The features of Cap analogs.
| Feature | Cap 0 Analogs | Cap 1 Analogs | Cap 2 Analogs |
| Methylation Pattern | m7GpppN (no ribose methylation) | m7GpppNm (first nucleotide methylated) | m7GpppNmNm (first two nucleotides methylated) |
| Immunogenicity | High (activates innate immunity) | Low (reduces immune detection) | Minimal (further immune evasion) |
| Translation Efficiency | Moderate | High | High to very high |
| Synthetic Complexity | Low | Moderate | High |
| Primary Application | Basic research, in vitro studies | Therapeutics, vaccines, gene therapy | Specialized high-performance applications |
Anti-reverse cap analogs (ARCAs) overcome the orientation issue of dinucleotide caps by incorporating a 3'-O-methyl group into the guanosine moiety, which sterically hinders the reverse orientation while leaving forward orientation unimpeded. ARCAs provide an important benefit as effectively all capped transcripts are translationally competent, which results in doubling protein expression over m7GpppG without increasing complexity of synthesis. ARCAs are still a Cap 0 cap analog, and thus remain a target for IFIT1 binding unless enzymatically methylated to a Cap 1; as such, they are largely used for translational biology and preclinical studies where orientation is more critical than immune evasion. The 3'-O-methyl substitution has been shown not to negatively affect binding to eIF4E, and the resulting mRNA exhibits increased stability against decapping enzymes, likely due to increased rigidity of the triphosphate bridge. Synthetic procedure for ARCAs is identical to that of standard caps, with the additional methylation step occurring on the guanosine precursor. The additional step adds a modest cost but does not affect scalability. The main utility of ARCAs is their ability to increase translational output without requiring changes to the IVT template, and thus represent an easy intermediate to both basic Cap 0 caps and more sophisticated co-transcriptional reagents. For clinical use, ARCA production will require qualification to demonstrate control of diastereomeric purity and lack of mutagenic impurities, which has thus far been an impediment for use in commercial vaccines.
Co-transcriptional capping reagents are trinucleotide or tetranucleotide analogs that directly install Cap 1 or Cap 2 structures during in-vitro transcription, avoiding the need for a separate post-transcriptional enzymatic modification step and achieving capping efficiencies of greater than 90%. The building block structure of these reagents is m7GpppNm–N, where the first nucleotide is pre-methylated at the 2'-position and the second nucleotide is the start site for transcription, forcing correct orientation so the nascent transcript is guaranteed to come off the IVT reaction with a mature, immune-evasive cap in place. Trinucleotide analogs can only be used for Cap 1 synthesis. If methylation is also pre-installed on the second nucleotide, a tetranucleotide reagent is created, and Cap 2 mRNA is generated in a single step. The primary advantage to this approach is process simplification: the single IVT reaction produces capped, translation-ready RNA, minimizing the number of steps, batch-to-batch variability, and total cost-of-goods. These reagents are ideal for GMP workflows because they are chemically defined materials that can be purchased from multiple qualified suppliers, which eliminates supply-chain risk. Performance is generally superior to dinucleotide caps and ARCAs in direct comparisons, with higher protein expression and lower immunogenicity. The primary disadvantage is template specificity: the DNA template sequence must begin with the matching initiation codon of the trinucleotide cap, which may require redesign of existing constructs. For new therapeutic programs, co-transcriptional capping is becoming the standard of care and has been adopted for rapid scale-up from research to commercial production.
Cap analogs are a type of chemical reagent with a wide variety of applications in basic science, vaccine development and therapeutic applications; overall they enable the synthesis of capped mRNA for a wide range of purposes. For basic molecular biology research, they are used to enable the synthesis of capped, translation-competent mRNA. In vaccine development, cap analogs are used to scale-up the synthesis of mRNA for a specific antigen (for example, a pathogen antigen for prophylactic vaccination, or a tumor neoantigen for therapeutic vaccination) as well as for therapeutic applications of mRNA such as protein replacement therapy, cancer immunotherapy, regenerative medicine and gene editing. In these applications, capped mRNA is delivered into cells where it serves as a template for the transient synthesis of either a therapeutic protein or gene editing machinery. Different types of cap analogs can be chosen to modulate specific properties of the mRNA, such as translational efficiency, stability and immunogenicity.
Cap analogs are also reagents commonly used in basic molecular biology research. In these studies, cap analogs are used to synthesize mRNA transcripts in vitro that are structurally and functionally similar to those found within cells. For example, cap analogs are often used in these types of experiments to create capped RNAs for translation in vitro, as a cap structure is necessary for ribosomal loading of the mRNA transcript. The ability to make capped transcripts without any cellular capping enzymes allows researchers to tightly control mRNA constituents to understand how changes to cap structure can affect translation, stability, or localization of the RNA transcript. Cap analogs are also used in research related to understanding cap-binding proteins like eukaryotic initiation factor 4E and its interactions in translation control. Additionally, cap analogs are used in mRNA decay studies, since the cap structure found at the 5' end of an mRNA transcript protects it from 5'-to-3' exonucleolytic decay while also being targeted by decapping enzymes. The chemical versatility of cap analogs also allow these reagents to be used in examining the structure-function relationship of this molecule. For instance, changes to the phosphate backbone, ribose, or base portions of cap analogs are used to study how these changes affect activity, which has been useful in understanding the molecular determinants for recognition of this structure by translation and innate immune sensors.
mRNA vaccines are another successful application of cap analog reagents. With these tools, large quantities of in vitro transcripts encoding immunogenic antigens, either viral or tumor-associated, can be rapidly produced. The choice of cap is important in vaccine design since it can affect the stability of mRNA both in formulation vehicles and once inside the cell, as well as its translational efficiency. Cap 1 and cap 2 are commonly used in vaccine design, as the addition of 2'-O-methyl modifications hide the transcript from being recognized as non-self RNA, thus preventing induction of innate immunity while improving protein expression. This contributes to the safety and efficacy of the final vaccine product, with the ability to induce both humoral and cellular immunity. The ability to rapidly generate capped mRNA using co-transcriptional capping using cap analogs dovetails with the manufacturing needs for vaccine production and allows the easy generation of large quantities of capped mRNA for inclusion in lipid nanoparticles. Cap analogs are used in therapeutic cancer vaccines which express patient-specific tumor neoantigens. In this context, the need for patient-specific, small-scale vaccine production with high batch-to-batch consistency makes cap analogs a practical and effective choice. The chemical diversity of these reagents also allows for the addition of stability-enhancing modifications that increase antigen expression, and with it, vaccine efficacy.
Cap analogs are also being used to develop mRNA-based therapeutics. In this context, they can be applied for vaccination, protein replacement, cancer immunotherapy, regenerative medicine, gene editing and other potential applications. Capped mRNA transcripts can be translated into proteins that can be used for protein replacement therapies. This can be used to compensate for protein deficiencies in patients with genetic diseases without permanently altering the genome. For applications like these, it is also important that the cap is optimized to allow for maximum translation with minimum immune response since this will allow repeated doses to be administered. Capped mRNA can also be used in cancer immunotherapy. Cytokines, co-stimulatory molecules, and chimeric antigen receptors can be expressed from capped mRNA. Capped mRNA can also be used to produce growth factors and differentiation signals in the context of regenerative medicine. It can also be used for gene editing, where capped mRNA can be used to express CRISPR-associated nucleases or base editors. This would be preferable to DNA-based expression vectors, since it would only express the editing machinery transiently and reduce off-target effects. Finally, features can be added to the cap analogs to improve targeting to specific tissues or cell types, which may be used to improve the therapeutic index of a treatment and reduce systemic exposure.
We offer a comprehensive range of cap analogs for mRNA synthesis designed to support reliable and efficient in vitro transcription (IVT) workflows. Our cap analog portfolio covers commonly used cap structures and capping strategies, enabling consistent 5' capping performance across a wide variety of mRNA research and production settings. Each product is developed with a focus on chemical quality, process compatibility, and reproducible results.
Our cap analogs are available in both research-grade and GMP-grade options, allowing users to select materials that align with their development stage and quality requirements. Research-grade cap analogs are well suited for basic mRNA studies, assay development, and process optimization, while GMP-grade materials support regulated mRNA production workflows that require enhanced quality control and documentation.
High capping efficiency and reproducibility depend on the quality of the cap analog. Our products are manufactured using tightly controlled processes to achieve high chemical purity and low impurity profiles, supporting consistent co-transcriptional capping and stable IVT performance. Rigorous quality control and analytical characterization ensure batch-to-batch consistency, helping minimize variability in mRNA yield and translation efficiency.
In addition to standard catalog products, we provide custom cap analog solutions to meet specific research or production needs. Customization options include alternative cap structures, tailored purity specifications, and formulation adjustments to ensure compatibility with unique IVT conditions. Our technical team works closely with customers to deliver cap analogs that integrate seamlessly into existing workflows and support long-term project requirements.
Selecting the right cap analog is a foundational step in achieving efficient mRNA synthesis, stability, and translation, and the optimal choice depends on your workflow, scale, and performance goals. Whether you are establishing a new IVT system or optimizing an existing process, our team is ready to support your mRNA capping strategy—contact us today to discuss your project and request technical information or a quotation.
References
Cap analogs are synthetic molecules used to introduce a functional 5' cap during IVT mRNA synthesis.
Cap analogs allow efficient and scalable capping during IVT, which is not possible with natural caps alone.
Most cap analogs are designed for co-transcriptional incorporation, though post-transcriptional methods also exist.
Yes, cap structure and orientation directly influence ribosome recruitment and translation initiation.
Most functional mRNA applications require proper capping to ensure stability and translation.

