ARCA caps were developed to prevent the in-vitro transcription artifacts of back incorporation that affect standard cap analogues. The addition of ARCA blocks the 3'-OH of the methylated guanosine, allowing the RNA polymerase to only start in one direction. This results in a higher percentage of translatable molecules and higher total protein yield, without changing downstream purification or immunogenicity characteristics.
ARCA is a second-generation cap structure which combines the immune-evading properties of Cap 1 methylation with a chemical property to prevent reverse incorporation. The result is a co-transcriptional capping reaction that produces >95% pure product, without needing post-synthesis enzymatic clean-up. The innovation involves a single atom difference – methylation or deoxygenation of the 3'-carbon of the guanosine ribose, such that the T7 RNA polymerase cannot start transcription in the reverse direction. This changes a probabilistic cap-mixing step into a deterministic high-yield step that is amenable to lyophilisation, microfluidic formulation, and other downstream manufacturing operations that are now common in mRNA production.
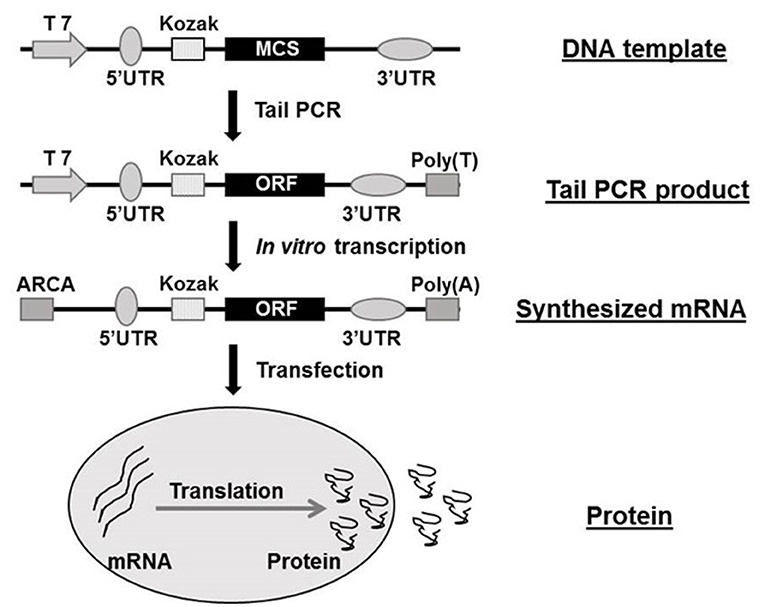 Fig. 1 Flow chart of mRNA synthesization in vitro1,5
Fig. 1 Flow chart of mRNA synthesization in vitro1,5
With standard in-vitro transcription, the RNA polymerase is capable of starting synthesis from either the 3'-OH or the 2'-OH of the incoming cap dinucleotide. This results in two transcript populations: the desired 5'-5' linked mRNA and a mirror-image species in which the cap is attached in the reverse orientation. The latter is translationally inactive due to steric interference by the reversed triphosphate bridge with eIF4E binding and rapid DXO cleavage, and it can account for up to a fifty percent loss of total protein product without inducing measurable innate immunity. The reverse incorporation is stochastic and cannot be reduced by manipulating cap-to-GTP ratios or reaction temperature; it is an unavoidable consequence of the symmetrical 3'-OH, 2'-OH geometry of the guanosine ribose. ARCA precludes this pathway by eliminating the 3'-OH, forcing the polymerase to start exclusively from the remaining 2'-OH, and ensuring that every transcript will have a self-recognized cap.
Native M7 GpppG dinucleotides have three major, inter-related limitations: heterogeneity of orientation, moderate affinity for eIF4E, and decapping sensitivity. The 3'-OH and 2'-OH groups are symmetric, allowing for T7 polymerase to start transcription both 5' and 3' to the G, creating a complex of properly and improperly capped RNAs that can be purified by costly enzymatic polishing or must be accepted with lower potency. In its properly oriented form, the native triphosphate bridge is a poor inhibitor of the interferon-inducible pyrophosphatase DXO, which cleaves the α-β phosphoanhydride bond and thereby tags RNA for 5'-to-3' exonucleolytic decay. Finally, the ribose hydroxyls hydrogen bond with eIF4E, but with much weaker affinity than the endogenous capped mRNAs, leading to suboptimal ribosome recruitment in the face of intracellular competition. All these limitations are amplified at scale, as batch-to-batch variation in the ratio of forward to reverse caps can lead to unacceptable levels of variability in protein expression, forcing developers to over-formulate (i.e., use higher molar dosing, driving up cost and reactogenicity).
ARCA was designed to turn cap orientation from a probabilistic liability into a fixed asset, to boost translational output without the need for post-transcriptional enzymatic capping or multi-step purification. Initial SAR work demonstrated that 3'-OH blocking with a methyl or deoxy group abolished reverse initiation while leaving the remaining 2'-OH intact as a T7 polymerase primer. Tetra- and pentaphosphate bridges were introduced in subsequent analogs to further increase intrinsic eIF4E affinity, and enhance ribosome recruitment. These bridged 2'-F analogs generate mRNAs that are translated as much as 2.5 times more efficiently than those capped with standard m⁷Gp₃G, without affecting immunogenicity or pharmacokinetic fate. ARCA-capped transcripts are also fully compatible with downstream transcript modifications such as N1-methylpseudouridine substitution or 3' poly(A) tail engineering, so developers can "stack" these optimizations without needing to re-optimize an entire synthesis workflow. The cost-benefit is not trivial: improved translational output means lower effective dose, lower lipid exposure, and simplified cold-chain handling and distribution, making ARCA an enabling technology for both pandemic-scale and personalized neo-antigen vaccines.
The ARCA architecture is defined by a single and well-positioned steric block (predominantly a 3'-O-methyl or a 3'-deoxygenation) in the N7-methylguanosine ribose, which abolishes the nucleophile for retro-priming but leaves the 5'-triphosphate exit vector unaltered; this minimal alteration locks initiation to one direction, does not compromise eIF4E recognition and provides a chemically inert scaffold for further phosphate or nucleobase decorations without affecting synthetic accessibility.
Reverse incorporation is due to the fact that the normal M7GpppG has two vicinal hydroxyls (2'-OH and 3'-OH) that both have equal opportunity to prime RNA polymerase. ARCA obviates this by pre-selecting the 3'-OH with either methylation or deoxygenation. Only the 2'-OH is then available for nucleophilic attack during initiation. This 1-atom difference eradicates the symmetry of the guanosine ring and results in a diastereomeric pair, which can be chromatographically separated to provide the pure "forward" isomer. The modification can also be used downstream to extend the phosphate tail and conjugate fluorophores, meaning the same orientation-control scaffold can be functionalized with handles without having to re-optimize polymerase kinetics. Crucially, the 3'-O-methyl group is inert and metabolically silent, so once the mRNA is translated the cap is cleaved by DXO and the resulting mononucleotide is processed through the normal nucleotide salvage pathways, rather than building up as a foreign metabolite that could initiate further toxicology evaluation.
Side-by-side comparisons show that ARCA results in an order of magnitude gain in translation without loss of metabolic camouflage. A regular M7 GpppG is equally divided between forward and reverse capped transcripts. Only the former can be translated and even those are quickly decapped, effectively a 50 % loss of working mRNA dose. ARCA circumvents this loss, producing a population where >95 % of the transcripts are translation-competent in the right orientation. This 2-3 fold increase in protein production per equal amount of RNA holds true across cell types and is additive to other optimization layers such as N1-methylpseudouridine (m¹Ψ) substitution and codon de-optimization. ARCA capped mRNA also exhibits modestly increased resistance to DXO, likely because the 3'-O-methyl group causes steric hindrance at the scissile phosphate. From a manufacturing perspective, ARCA requires the same one-pot co-transcriptional addition as regular caps, but avoids the price tag and batch-to-batch variability of post-transcriptional enzymatic capping, while still leaving the option to upgrade to Cap 1 via a separate methylation step for additional immune stealth.
This orientation control is co-transcriptional, and is achieved by providing ARCA at molar excess to GTP, such that the polymerase more likely adds the modified cap than the naturally-occurring nucleotide. Because only one, the 2'-OH, is present in ARCA, each initiation event results in a transcript with a 5' end of known orientation for eIF4E binding. The reaction is rapid and works under normal IVT conditions (37 °C, pH 8, mM NTPs), and does not require proprietary buffer systems or lengthy incubations. To ensure the correct competitive binding between ARCA and GTP, the cap: GTP ratio is adjusted and a high-affinity T7 variant can be used that is only slightly more discriminatory against GTP to bias the capping efficiency to the upper end of the 60 to 80% range observed for ARCA reactions. Orientation purity can be checked by digestion with nucleases followed by RP-HPLC, which separates forward and reverse caps based on differing retention times. The same method can be used to confirm that ARCA is not present in the final drug substance. Since all caps are of the correct orientation there is no need for enzymatic re-capping as a downstream processing step.
Table 1 Comparison of cap platforms
| Attribute | Standard M7 GpppG | ARCA (m²⁷,³'-OGp₄G) | Clinical implication |
| Reverse incorporation | Up to 50 % | < 5 % | Higher yield per batch |
| eIF4E affinity | Moderate | High | Lower effective dose |
| DXO resistance | Low | Moderate | Longer half-life |
| Manufacturing | Two-step polishing | One-step co-transcriptional | Lower COGS |
| Regulatory status | Well-established | Compendial-grade available | Accelerated CMC |
ARCA increases protein expression by transforming the probabilistic and error-prone capping reaction into a deterministic process in which every transcript is capped with a correctly oriented, structurally stable cap optimized for ribosome recruitment. This is accomplished without any changes to the coding sequence or delivery vehicle, as the analog itself enforces orientation purity, resists premature decapping, and provides a high-affinity landing platform for the eIF4E complex to drive faster initiation and a lower fraction of mRNA targeted for cellular quality-control pathways.
Chemically conventional M7 GpppG, in contrast, gives rise to a near-equimolar mixture of forward and reverse caps. This is because T7 RNA polymerase is capable of initiating from both the 2'-OH or 3'-OH of the incoming dinucleotide. Reverse-oriented caps are inert for translation: they do not recruit eIF4E, are rapidly cleaved by DXO, and can act as effective decoys that dilute functional transcripts. ARCA removes this inefficiency by blocking the 3'-OH with a methyl or deoxy group, so that the polymerase must prime exclusively from the 2'-OH. The result is a population in which >95 % of transcripts carry a cap that is correctly oriented for ribosome binding, translating into a two- to three-fold increase in protein expression at equivalent RNA input without increasing lipid exposure or immunogenicity. The improvement is seen across cell types and is additive with other optimization layers such as N1-methylpseudouridine substitution, codon de-optimization, and UTR engineering, so orientation control is a foundational rather than marginal enhancement.
ARCA-capped mRNA has a cap surface that is structurally more similar to endogenous transcripts than chemically capped mRNA, and eIF4E binding is tighter and assembly of the 43S pre-initiation complex is faster. Addition of extended phosphate backbones (tetra- or pentaphosphate) commonly incorporated into the ARCA structure further boosts the intrinsic affinity for the cap-binding complex, with no change in the basic orientation control element, in effect "super-charging" the recruitment step. The result is a more rapid transition from initiation to elongation and a smaller time window for the transcript to be decapped or cleaved by endonucleases. In ribosome profiling experiments, ARCA-capped mRNA had higher ribosome density over the first fifty codons, consistent with more efficient initiation and less drop-off at upstream open reading frames. The effect is magnified under stress conditions where there is competition for limiting eIF4E; while ARCA-capped transcripts remain bound to polysomes, conventional capped mRNA is displaced, allowing continued protein production under interferon-rich or nutritionally depleted conditions.
Head-to-head studies in several mammalian cell lines, primary dendritic cells, and in-vivo mouse models confirm that ARCA provides a 2-3 fold increase in reporter protein output compared to traditional M7 GpppG, and this benefit holds across electroporation, lipid nanoparticle and naked RNA delivery technologies. This is not just a technical point: mRNA encoding influenza hemagglutinin with an ARCA cap drives higher antibody titers and greater neutralization breadth at a lower dose than conventionally capped mRNA, showing that the gain at the translational level leads to a real immunological benefit. Furthermore, the correctly oriented cap has also been shown to be modestly more resistant to DXO-mediated decapping, which slightly prolongs its half-life in a cytosolic extract and does not lead to any novel metabolites that might require additional toxicology evaluation. Critically, this gain is maintained when ARCA is used with CleanCap or enzymatic Cap-1 methylation, suggesting that orientation control and 2'-O-methylation are orthogonal improvements that can be combined without compromising either. Overall, this makes ARCA a drop-in replacement that can improve both potency and manufacturability, positioning it as the default choice for next generation mRNA therapeutics.
ARCA's orientation-based capping chemistry has moved synthetic mRNA from a laboratory reagent to a precision therapeutic platform, with every transcript guaranteed to be translationally competent, reducing effective doses, shortening development timelines, and allowing for direct comparison of delivery platforms without the variable of cap heterogeneity. ARCA has been integrated into basic research, vaccine development, and advanced cell-engineering workflows where transient but high-level protein expression is required.
Table 2 Map of ARCA applications
| Application area | Primary challenge | ARCA contribution | Typical downstream benefit |
| Protein expression studies | Reverse-cap noise | >95 % forward orientation | Clean quantification of UTR/codon effects |
| Vaccine platforms | Low antigen yield | 2–3× higher expression | Dose-sparing, reduced reactogenicity |
| Difficult proteins | Rapid degradation | Sustained translation window | Achieves functional concentration before clearance |
ARCA-capped mRNA has also become common in basic research applications that attempt to understand the relationship between the delivered transcript and the final protein yield; the homogeneity of the forward cap obviates the stochastic failure of a reverse-cap reaction as a complicating factor. Dual-fluorescence reporters with ARCA-capped EGFP mRNA and an orthogonal Cy5 label for RNA localisation are one example, allowing the normalisation of the reporter signal to the true intracellular mRNA abundance to determine vector-specific rate-limiting steps (i.e. endosomal entrapment, early degradation etc.). The stability of the cap during microfluidic mixing and nebulisation make it useful in mechanistic studies that attempt to parse out the kinetics of endosomal release from translation initiation. The fact that ARCA can be added co-transcriptionally along with modified nucleotides such as 5-methoxy-CTP or pseudouridine also allows for the concurrent reduction of innate immune activation while boosting expression. This can provide a "clean" background to evaluate other effects such as codon usage, UTR strength or RNA-binding proteins without cap-related confounders.
For both prophylactic and therapeutic vaccines, ARCA provides an economical approach to high-titer antigen expression by optimizing the proportion of transcripts translated to immunologically relevant peptides. The co-transcriptional incorporation allows for ~80 % capping efficiency in a single 30-minute reaction, without the need for post-synthetic enzymatic polishing or extension of the manufacturing timeline. The resulting mRNA is fully compatible with downstream modifications such as lipid nanoparticle encapsulation, lyophilization and intradermal or intramuscular injection while providing consistent seroconversion at lower RNA doses than conventionally capped material. In addition to infectious-disease applications, ARCA is now being applied to personalized cancer vaccines where tumor neoantigen libraries must be produced in short timeframes; the cap uniformity ensures that each antigen is expressed at similar levels, eliminating the under-representation of epitopes that could otherwise allow immune escape.
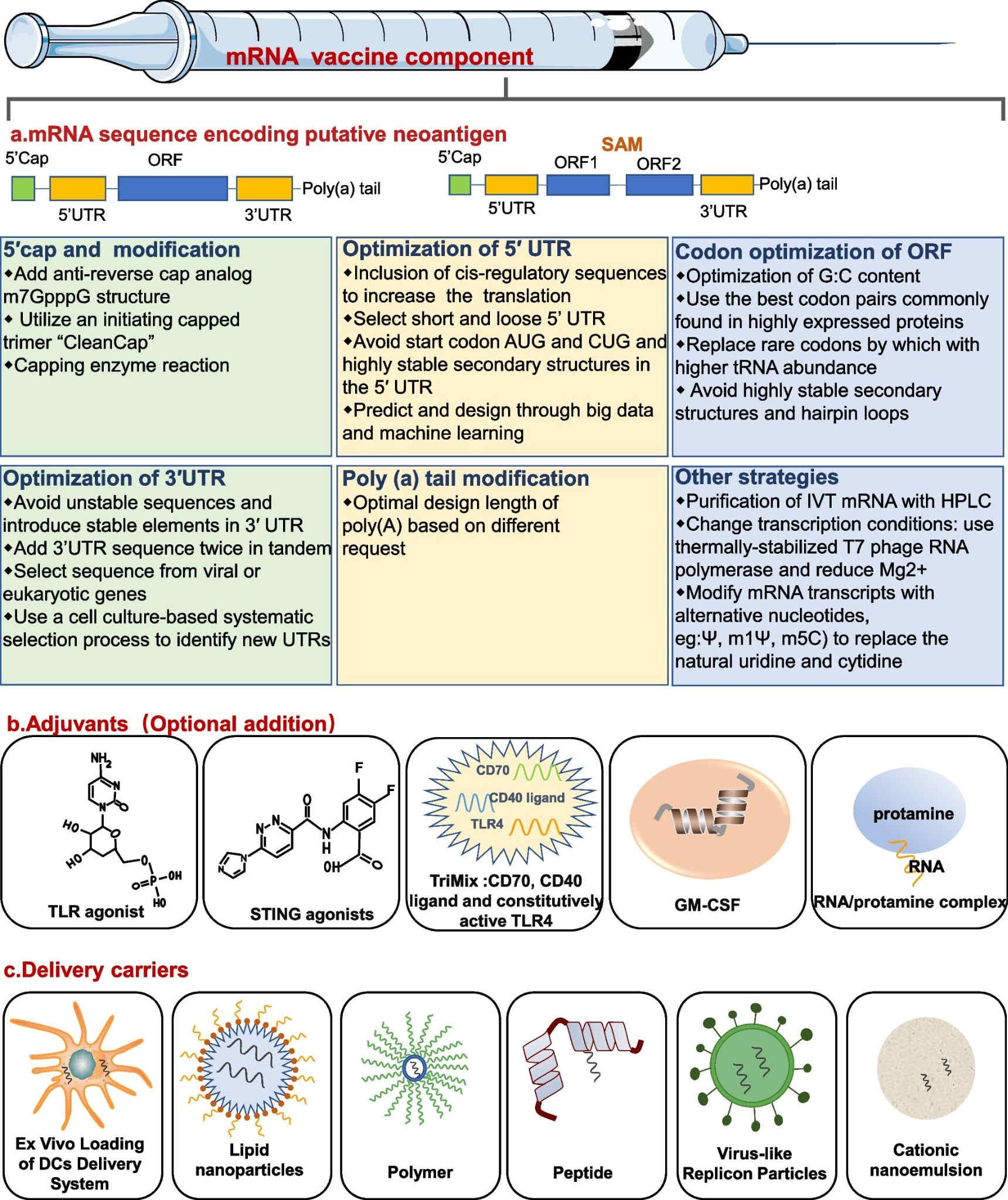 Fig. 2 The main components of mRNA tumor vaccine2,5
Fig. 2 The main components of mRNA tumor vaccine2,5
ARCA is of particular utility for the expression of proteins that are toxic, aggregation-prone, or subject to rapid proteasomal degradation, as the increased translational yield allows the desired intracellular concentration to be reached prior to protein clearance. In differentiation protocols with induced-pluripotent stem cells, ARCA-capped mRNA encoding mutant transcription factors leads to sustained nuclear accumulation without repeated transfection to accelerate lineage commitment while minimizing cytotoxicity. The stability of the cap under repeated electroporation also permits sequential delivery of multiple smRNAs (each capped with ARCA) to choreograph complex differentiation trajectories (e.g. oligodendrocyte maturation or cardiomyocyte specification). In genome-editing applications, ARCA-capped Cas9 or base-editor mRNA has sufficient ribosome engagement time to achieve biallelic editing prior to transcript degradation, which decreases the need for high-multiplicity transfection that can initiate p53-mediated stress responses. In sum, these use-cases suggest ARCA will be an enabling reagent for any application where transient but robust protein expression is the bottleneck to success.
Selection among ARCA, enzymatic Cap 1 and CleanCap-type trinucleotides is a risk/reward decision matrix of translational potency vs immunogenicity risk, manufacturing complexity and supply-chain assurance. ARCA trinucleotides provide >95 % assurance of forward orientation, but are still Cap-0 and IFIT1-activating, so are limited to adjuvanted contexts. Enzymatic Cap 1 has the advantage of true "self" methylation, but adds an additional enzymatic step with attendant batch-to-batch variation. CleanCap trinucleotides provide Cap-1 as a part of the co-transcriptional synthesis process, with >90 % efficiency, no competition with GTP and are already in place in billion-dose supply chains and hence are by default the choice for clinical programmes prioritizing speed, yield, and reactogenicity control.
CleanCap consistently yields a higher fraction of translationally competent transcripts in a single IVT step when compared to either ARCA or post-transcriptional Cap 1. While ARCA may have a comparable forward yield of ~80 %, its Cap-0 identity is not able to escape a checkpoint in the translation initiation machinery which stalls ribosomes bound to ARCA-capped mRNA by virtue of their recruitment of IFIT1. In contrast, CleanCap's intrinsic Cap-1 methylation enables cap-dependent translation initiation to double reporter gene output relative to ARCA at the same total RNA mass, independent of other features. Enzymatic Cap 1 can achieve comparable fidelity in vitro, but only after additional methylation and purification steps that decrease yield. The trinucleotide design also controls start-site fidelity, forcing an AG dinucleotide with CleanCap and minimizing AUGs immediately upstream which would otherwise be translated into truncated products. In different cell lines and in-vivo models, CleanCap-capped mRNA is seen to have higher polysome loading and lower decapping rates, resulting in lower effective doses and decreased lipid exposure for the same pharmacodynamic endpoint.
ARCA-capped mRNA is targeted as foreign due to the lack of 2'-O-methylation of the first nucleotide. This leads to the activation of IFIT1, triggering the interferon response, which can attenuate vaccine immunogenicity or induce a febrile response. CleanCap analogs incorporate this modification co-transcriptionally and have a Cap-1 signature that is not recognized as foreign, thereby mitigating systemic cytokine release without the need for a separate enzyme step. Post-transcriptional Cap 1 circumvents this issue, but adds the potential for incomplete methylation if enzyme activity drifts, which is a poorly controlled variable that is under increasing scrutiny from regulators within the scope of ICH Q6B. For therapeutic proteins where repeat dosing is expected, the innate immune silence from CleanCap is believed to be required to prevent anti-drug antibody formation or cumulative interferon toxicity.
CleanCap simplifies the IVT workflow into a one step closed-tube reaction which requires no post-capping purification. This cuts processing time in half and reduces the risk of open-handling contamination. ARCA requires only a change in cap analog but is still a dinucleotide that competes with GTP. It must be used with careful optimization of NTP ratios and typically results in a lower overall RNA yield. Enzymatic Cap 1 requires two additional steps: incubation with a methyltransferase and purification to remove the enzyme after reaction. Each of these steps incurs additional cost and increases analytical burden. On a cost-per-functional-transcript basis, CleanCap is usually lower than enzymatic Cap 1 once yield losses and QC overhead is factored in, while ARCA remains the least expensive for research grade material where immune activation is acceptable.
ARCA is still preferred when speed and low-cost of synthesis are the dominant factors, and some level of innate immune activation is acceptable or even desired. In oncology vaccine studies where interferon signalling is used for DC maturation, or in basic-science screens where cap structure is not the experimental variable, ARCA's single-addition workflow and large vendor selection are attractive. For personalized neo-antigen panels that require dozens of constructs to be made in parallel within days, the lower unit price and on-hand availability of ARCA will outweigh the performance hit. However, as a candidate approaches pre-clinical toxicology or clinical manufacture, the immunogenicity and yield advantages of CleanCap-type analogs will generally outweigh the cost difference, making ARCA a useful stepping-stone to more refined cap structures.
We offer a comprehensive range of anti-reverse cap analogs (ARCA) designed to improve cap orientation and translation efficiency in in vitro transcribed (IVT) mRNA. Our ARCA products are developed to address the limitations of conventional cap analogs by minimizing reverse cap incorporation, resulting in a higher proportion of correctly capped mRNA and more efficient protein expression. From early-stage research to clinical development, our ARCA solutions are manufactured with a strong focus on quality, consistency, and compatibility with established IVT workflows.
Our high-purity ARCA products are optimized for research and preclinical mRNA studies, where reliable translation efficiency and experimental reproducibility are critical. These ARCA cap analogs support efficient co-transcriptional incorporation during IVT and help maximize the fraction of correctly oriented caps, leading to improved ribosome recognition and higher protein expression levels. They are well suited for mechanistic studies, reporter assays, and early-stage mRNA optimization, providing consistent performance across commonly used IVT systems.
For regulated applications, we offer GMP-grade ARCA manufactured under controlled conditions to meet the quality and documentation requirements of commercial mRNA development. These products are suitable for use in mRNA vaccines, therapeutic mRNA, and gene therapy programs, where cap structure and orientation directly affect efficacy and safety. GMP-grade ARCA is supplied with comprehensive quality documentation to support process validation, regulatory submissions, and long-term manufacturing programs.
Consistent translation performance requires cap analogs with stable and reproducible quality. Our ARCA products undergo rigorous quality control and analytical characterization to ensure consistent chemical identity, purity, and functional performance. This focus on batch-to-batch reproducibility supports reliable IVT mRNA synthesis, minimizes variability in translation efficiency, and helps maintain consistent protein expression during process development and scale-up.
In addition to standard catalog products, we provide bulk supply and custom ARCA solutions to support large-scale manufacturing and specialized process requirements. Custom options include tailored specifications, alternative formulations, and flexible packaging to align with customer-specific IVT workflows. Our scalable manufacturing capabilities and supply chain reliability ensure uninterrupted access to ARCA materials for both short-term projects and long-term development programs.
Optimizing cap orientation is a proven strategy for enhancing mRNA translation efficiency and protein expression, and selecting the right ARCA can make a measurable difference in IVT performance. Whether you are optimizing research-scale experiments or advancing clinical mRNA programs, our team is ready to help you identify the most suitable anti-reverse cap analog solution—contact us today to discuss your project and request technical support or a quotation.
References
ARCA prevent reverse cap incorporation during IVT, ensuring correct cap orientation for translation.
Reverse-oriented caps cannot be recognized by translation initiation factors, reducing translation efficiency.
By increasing the proportion of correctly oriented caps, ARCA enhances ribosome binding and translation initiation.
Yes, ARCA is designed to work with commonly used RNA polymerases and IVT workflows.
ARCA is recommended when maximizing translation efficiency and reducing variability are critical.

