Nucleoside monomers are the programmable alphabet of gene medicines: their heterocycles encode sequence identity, their sugars dictate helical geometry and metabolic half-life, and their phosphates provide conjugation handles for tissue-targeting ligands; whether the final drug is an siRNA duplex, an mRNA vaccine or a gapmer antisense, therapeutic outcome must first be written with chemically disguised yet Watson–Crick-readable nucleosides that retain affinity while evading innate sensors.
Nucleoside monomers are interesting for a number of reasons: they are the only molecular pixels to straddle biology's informational layer and medicinal chemistry's manufacturing layer: their 2'-OH/2'-H toggle chooses RNAi or RNase-H pathways; their base edits (Ψ, m5C, 8-aza-G) silence TLR3/7/8 and RIG-I; their lipid or GalNAc conjugates influence biodistribution; that one choice leads to a divergence in synthesis chemistries, impurity limits and regulatory filing categories, making monomer choice the first irreversible engineering decision which ultimately dictates whether an oligonucleotide will survive circulation, enter cells and engage its target safely.
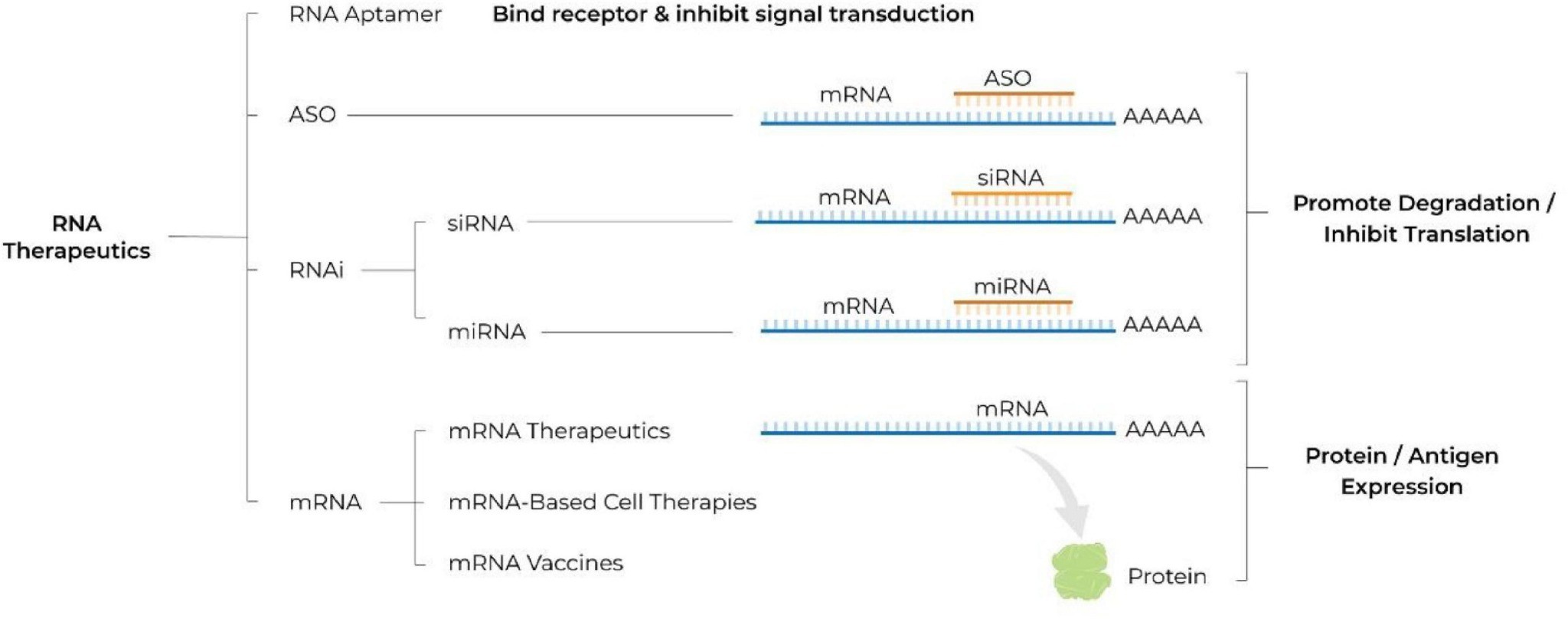 Fig. 1 Schematic illustrating different classes of RNA therapeutics.1,5
Fig. 1 Schematic illustrating different classes of RNA therapeutics.1,5
Nucleoside monomers are an indispensable first step in every clinically developed oligonucleotide platform. In siRNA, 2'-fluoro-uridine and 2'-O-methyl-cytidine are incorporated within the guide seed to rigidify the strand against serum nucleases but maintain A-form geometry for argonaute loading; N1-methyl-pseudouridine is used in non-seed positions to disrupt TLR7/8 recognition but not reduce catalytic turnover. In mRNA, full replacement with N1-methyl-pseudouridine and 5-methyl-cytidine abrogates PKR and RIG-I signalling, so that self-amplifying replicons can express monoclonal antibodies at microgram-per-millilitre titres in serum. In gapmer antisense, central tracts of deoxyguanosine recruit RNase H1 to cleave huntingtin or SMN2 transcripts, while flanking 2'-MOE-cytidine wings improve affinity and block exonucleases. In this way, nucleoside monomers serve as addressable chemical pixels to toggle catalytic, translational or degradative phenotypes on demand.
Table 1 Monomer Edit vs Functional Gain
| Edit | Functional Gain | Platform Use |
| 2'-OMe | ↑ Tm, ↓ RNase H | siRNA guide |
| 5-Me-C | ↓ TLR7 | mRNA cap |
| Ψ | ↑ stability | mRNA body |
The modalities covered by the therapeutic oligonucleotide portfolio have broadened far beyond the traditional gapmer antisense domain to include self-amplifying mRNA, circular RNA, bispecific aptamers and CRISPR guide RNAs, each with their own unique nucleoside requirements. Fast kilobase synthesis of self-amplifying mRNA can only be achieved with polymerase-mediated Ψ and m5C incorporation, while circular RNA vaccines are armed with 2-amino-6-thio-guanosine to facilitate disulfide crosslinks that redirect ribosomes to shunt around the molecule before it is cleaved. Bispecific aptamers incorporate 8-bromo-guanosine to pre-select for syn conformation, driving the formation of parallel G-quadruplexes that bind von Willebrand factor with nanomolar affinity. Emerging modalities such as these are starting to treat the nucleoside monomers themselves as a series of structural pixels, where bromo tunes topology, thio triggers redox cleavage and propynyl locks down the lattice, enabling entirely new geometries of therapeutic beyond traditional duplex or quadruplex scaffolds.
The choice of monomer also strongly influences the overall production strategy: Ψ or m1Ψ triphosphates are produced in aqueous solution, need ion-exchange purification, freeze-drying and a −20 °C cold-chain, increasing variable cost and depreciation of capital equipment. Crystalline 2'-OMe-cytidine phosphoramidites, on the other hand, are "middle-sized" organic molecules that crystallize from ethanol–water and remain stable in years under ambient humidity. The product loss profiles are different as well: the solid-phase synthesis of gapmers with 5-Me-C in the core will result in linear loss of yield, while enzymatic mRNA synthesis with Ψ will show exponential decline after 4 kb and therefore also requires downstream RNase III polishing and tangential-flow concentration. Regulatory authorities now require safety data of every metabolite of modified nucleosides, driving developers to "biodegradable" modifications, which would be converted to natural nucleosides upon catabolism. The monomer choice therefore maps to differences in facility design, environmental footprint and commercial potential. This choice is the kinetic bottleneck which decides if an oligonucleotide programme can flow from discovery to late-stage GMP manufacture without re-synthesis or specification drift.
siRNA, mRNA and antisense oligonucleotides (ASOs) represent the 3 classes of therapeutic nucleic acids. However, they use different 'languages' to operate. siRNA provides dsRNA that is loaded into RISC to cleave the target mRNA. mRNA is a single-stranded transcript that is translated into protein in the cytosol. ASOs are single-stranded DNA/RNA hybrids that either recruit RNase H or sterically block splice sites. The combination of different mechanisms, lengths and chemistries requires developers to hard code potency, stability and immune evasion into the monomer units, rather than relying on post-synthetic rescue. This also allows the same nucleoside toolbox to be used to develop all 3 modalities without cross-contamination or redundant validation.
Table 2 Molecular and manufacturing dichotomy across platforms
| Platform | Strand | Length (nt) | Key monomer edit | Chemistry route | Functional read-out |
| siRNA | dsRNA | 19–23 | 2'-F-U/C | Solid-phase amidite | RISC cleavage |
| mRNA | ssRNA | 500–5000 | Ψ-U / 5-Me-C | Enzymatic IVT | Protein expression |
| ASO | ssDNA/RNA | 16–20 | 5-Me-C / LNA-G | Solid-phase amidite | RNase H cleavage |
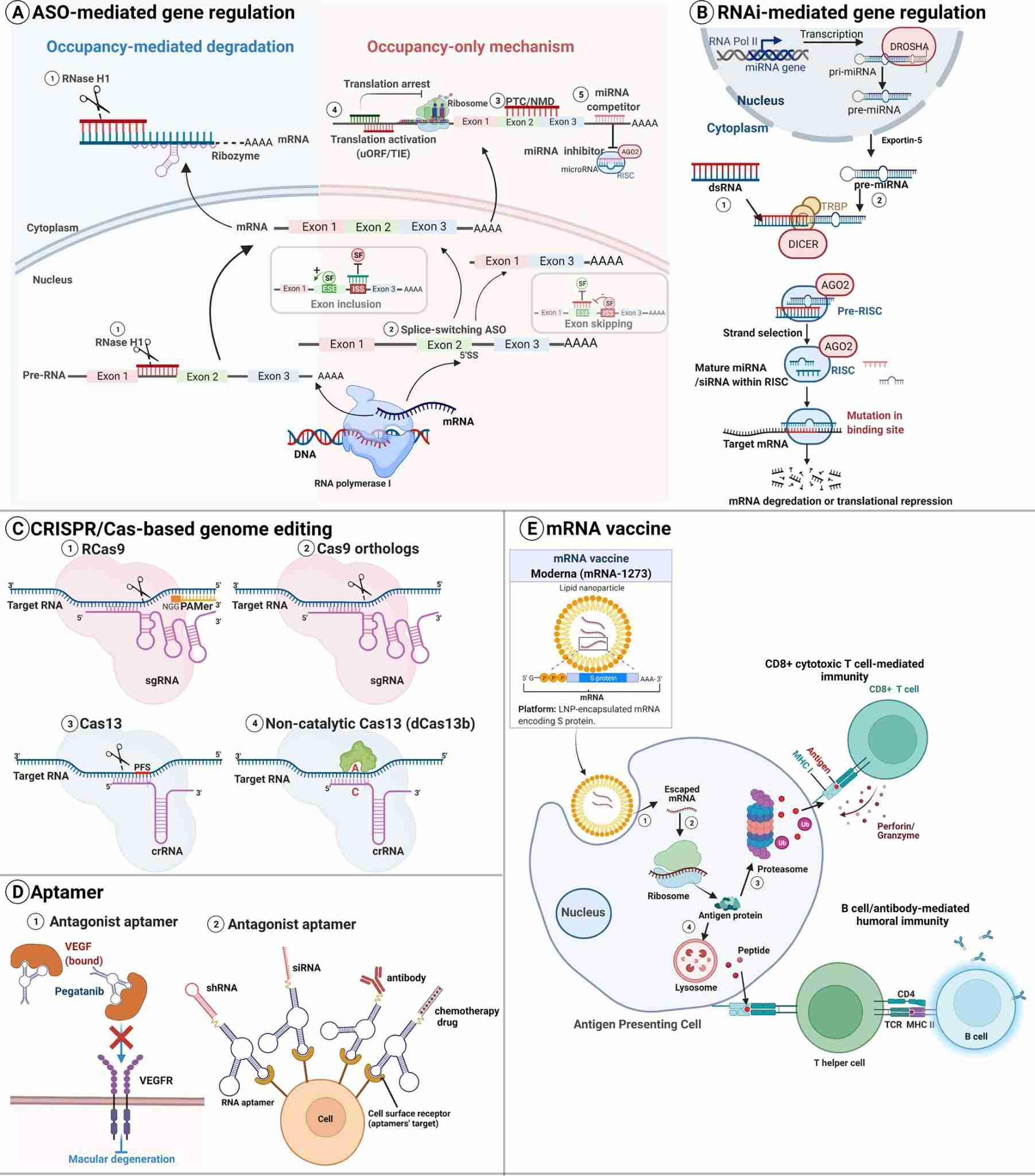 Fig.2 Types of RNA-based therapeutics and modes of action.2,5
Fig.2 Types of RNA-based therapeutics and modes of action.2,5
siRNA is 19–23 nucleotide-long double-stranded RNA which exploits the endogenous RNA interference pathway. After entering the cell, the guide strand is loaded into the RISC and the passenger strand is discarded; if the guide strand is perfectly complementary to the target mRNA, Ago2-mediated endonucleolytic cleavage is followed by exonucleolytic degradation of the cleaved mRNA fragment, and the gene is silenced without modifying the underlying DNA sequence. Because the same duplex can be chemically edited (2'-fluoro-uridine, 2'-O-methyl-cytidine) to increase serum half-life and to reduce off-target breathing, developers encode potency and safety at the monomer level, so that the same solid-phase amidite run results in stereopure siRNA whose pharmacokinetics are pre-encoded prior to scale-up.
Antisense oligonucleotides (ASOs) are single-stranded, 16–20 nucleotide, DNA/RNA chimeras that hybridize to complementary RNA through Watson–Crick base pairing. The central DNA "gap" recruits RNase H to cleave the target transcript, while 2'-O-methyl or locked nucleic acid (LNA) "wings" increase binding affinity and confer nuclease resistance. Since the same sequence can be designed to block a splice site or modulate poly-A selection, ASOs can be switched between gene knock-down and isoform modulation simply by changing the location of the bases, allowing the same crystalline amidite feed to produce both catalytic cleavage and splice correction without any post-synthetic rescue.
mRNA is a 500–5000 nucleotide, single stranded transcript that is translated to protein in the cytosol; introduction of N1-methyl-pseudouridine and 5-methyl-cytidine during the triphosphate stage reduces RIG-I and TLR7/8 activation and increases ribosomal decoding speed. Because the same transcript can be capped, poly-adenylated and encapsulated into lipid nanoparticles during the same IVT run, manufacturers pre-encode both immune evasion and translational potency at the monomer level. This guarantees that the same enzymatic reactor will make self-amplifying or circular mRNA whose pharmacokinetics are pre-encoded before encapsulation even begins.
siRNA duplex is 19–23 nt, acts by RISC-dependent cleavage; ASO single strand is 16–20 nt, acts by RNase H recruitment or steric block; mRNA single strand is 500–5000 nt, acts by ribosome-mediated translation. The three lengths are 2 orders of magnitude apart and thus requires careful consideration of base-editing densities, where e.g. siRNA requires 2'-F and 2'-OMe wings for seed binding, ASO contains 5-methyl-C for immunomasking, and mRNA contains Ψ-U and 5-Me-C for translational enhancement—all added to the solution as the same crystalline amidite or triphosphate feed, thus bringing all three platforms under a single nucleoside toolbox without risk of cross-contamination or duplicate GMP validations.
The programmable alphabet of siRNA is its nucleoside monomers: their ribose-base conjugates can be locked, fluorinated or N-alkylated at the monomer stage to encode seed-region rigidity, duplex stability and serum half-life without altering the universal amidite coupling cycle—thereby allowing the same solid-phase synthesiser to deliver 19–23 nucleotide double-stranded RNA whose guide-strand potency, passenger-strand disposal and off-target rejection are pre-encoded at the nucleoside level before scale-up begins.
siRNA duplexes make use of thermodynamic asymmetry: only the guide strand should have a lower melting temperature in the 5'-end, to enforce loading of the correct strand in RISC and therefore discard of the passenger strand. Placement of 2'-fluoro-uridine in positions 1, 7 and 14 on the guide strand decreases the local Tm and speed up Ago2 anchoring. Addition of 2'-O-methyl-cytidine on the passenger strand increases rigidity and decrease potential off-target hybridisation. As both of these edits can be introduced as regular amidites, one solid-phase run delivers both catalytic potency and metabolic stability in one vial, making sure the strand asymmetry is encoded at the monomer level rather than being imposed by post-synthetic annealing.
2-thio-uridine enhances base-stacking and seed-region rigidity without affecting Argonaute loading, while locked-cytidine (LNA-C) raises melting temperature and increases serum half-life. As these sugar and base edits are completely orthogonal to standard amidite chemistry, developers can stack 2-thio-U on top of 2'-OMe-C to build third-generation duplexes that offer both high-affinity recognition and nuclease resistance in a manner that tunes silencing efficiency during synthesis rather than downstream formulation.
C-5 propynyl-uridine enhances π-overlap and increases Tm by ~2 °C per residue, thereby tightening guide-target binding and reducing seed-region breathing; pseudouridine flattens the base and restores silencing activity lost by central mismatches. As these base edits are introduced at the monomer stage, the same duplex presents both high-affinity anchoring and mismatch discrimination, ensuring that target recognition is encoded at the nucleoside level rather than imposed by backbone swaps.
siRNA monomers are synthesised as crystalline amidites: 2'-fluoro-uridine and 2'-O-methyl-cytidine are protected with standard DMT and acyl groups that are stable under anhydrous tetrazole activation to give ≥98 % coupling efficiency. With no hydrolysis of the same amidite for ≥12 months at −20 °C, a single master batch can support multiple siRNA programmes thus collapsing inventory complexity, so that the final duplex behaves as a single molecular entity instead of a heterogeneous mixture.
Nucleoside monomers are the fundamental "letters" that encode the sequence specificity and mechanism of action of antisense oligonucleotides. The base-sugar identity (A, T/U, C, G) of each monomer dictates Watson-Crick pairing, while the chemical modification (2'-F, MOE, LNA, phosphorothioate) of each monomer determines serum stability, RNase H recruitment and binding affinity. The choice of DNA vs RNA monomers, sugar pucker, and backbone chemistry in gapmer or steric-block designs are all made at the level of the monomer. Thus raw-material selection is the first major control point for clinical potency, safety and manufacturability.
RNase H–competent gapmers incorporate a central "gap" of 2'-deoxy nucleosides to form a DNA-RNA heteroduplex and recruit RNase H1 while the flanking domains are modified with 2'-O-MOE or LNA ribosides to increase the Tm of the oligonucleotide and provide exonuclease protection. Steric-block ASOs completely forego the use of DNA monomers and are composed of tightly binding 2'-O-MOE, 2'-F, or LNA ribosides that do not promote RNA cleavage. In this way, monomer choice dictates mechanism a priori (DNA to degrade, RNA-like to block).
2'-O-MOE increases stacking energy and lipophilicity,-O-MOE or 2'-MOE-C derivatives can be used which are nuclease-resistant but do not recruit RNase-H, so that cleavage takes place only at the target RNA. Steric-blocking ASOs (splice-switchers) avoid DNA completely; they are synthesised as a mosaic assembled from 2'-O-Me, increasing Tm ~2 °C per modification and greatly facilitating RNase-H recruitment if they are in wings. 2'-F forces the sugar into a C3'-endo pucker, increasing hybridisation energy without increasing steric bulk, so it is useful for splice-switching or as an anti-miR 2'-F or locked nucleic acid (LNA) amidite to increase Tm and sterically block spliceosome access without cleavage of the RNA. As the decision is encoded in the monomer feedstock, the same synthesiser can switch from gapmer to splice-switcher merely by exchanging amidite bottles, mapping chemical diversity directly to mechanism-specific design.
Phosphorothioate linkages (added by sulfurizing the phosphite after each coupling) are built in at the monomer level and have profound effects on biodistribution: high plasma protein binding, low renal clearance, and days-long half-lives (vs. minutes). Sugar modifications can affect uptake as well: fully 2'-O-methyl gapmers are taken up by liver and kidney via scavenger receptors, whereas LNA-rich sequences partition into muscle and tumor tissue. In this way, the monomer recipe determines not just affinity, but also which organs will have the highest drug levels.
Chemical purity of GMP-grade phosphoramidites is typically ≥99.5 %, with <0.2 % water, <5 ppm total metals and ≥99 % β-anomer content; any non-compliance propagates as n-1/n+1 impurities or depurination sites in the drug substance. Batch-to-batch consistency is ensured by ¹H-NMR, LC-MS and chiral HPLC, and forced-degradation studies are conducted to demonstrate that no new genotoxic impurities form upon storage of the drug substance at 40 °C/75 % RH. Because ASOs are single-stranded, even a 0.5 % α-anomer impurity can decrease Tm and activate innate immunity, so anomeric purity is tracked as rigorously as chemical purity throughout the development program.
Nucleoside monomers are the "letters" that spell out mRNA stability, immunogenicity, and translational potency. In IVT, each monomer is supplied as a 5'-triphosphate ribonucleoside, with its base identity (A, U, C, G) encoding the message and its chemical modification (Ψ, m1Ψ, m5C) silencing innate sensors and tuning secondary structure. Modifications are introduced at the monomer level—not post-synthetically—traveling intact through polymerase, capping enzyme, and purification. The result is every codon carrying the desired chemical signature from the first base to the poly(A) tail.
Monomer usage directly impacts codon choice and decoding kinetics. Replacing uridine with Ψ or m1Ψ can eliminate U-rich sequences that cause ribosomal stalling and activation of PKR, while not disrupting wobble-pairing accuracy. In addition, the use of synonymous codons, whose cognate tRNAs are more abundant in the host cytosol, can eliminate rare-codon clusters to speed up elongation kinetics and minimize frameshifting. Such monomer-level codon optimization is implemented during IVT template design so that the final mRNA contains chemically modified, but codon-optimized, codons without the need for post-transcriptional editing.
Ψ and m1Ψ were added as full uridine replacements during IVT. Both can Watson-Crick pair without being detected by TLR7/8 and RIG-I. m1Ψ also inhibited PKR, which was preventing eIF2α phosphorylation and loss of cap-dependent translation. The other monomers m5C, m6A, and s2U were added at much lower levels to help adjust secondary structure without loss of decoding accuracy. Addition of these at the level of triphosphate monomers also meant that the polymerase added them evenly, and the immunoevasiveness was encoded from the first codon to the poly(A) tail.
Ψ and m1Ψ elevate Tm by ~1 °C per residue, resulting in more rigid secondary structure and consequent resistance to RNase L cleavage. More compact structure also impedes deadenylation and decapping, with net effects on functional half-life in vivo increasing from minutes to hours. Ψ-rich UTRs also form fewer stable stem-loops, enhancing eIF4A-mediated unwinding and thus increasing initiation rate. The choice of monomer at IVT stage thus predetermines both kinetic stability of the message and rate at which ribosomes decode it; neither effect can be undone by later formulation.
Enzymatic synthesis is based on recombinant polymerases and NTPs, and the modified base must be accepted by the enzyme. This limits modified bases to m¹Ψ, 5-methyl-C and N6-methyl-A routinely. Chemical synthesis uses phosphoramidite monomers on solid-phase resins. Chemical synthesis allows broader modification (2-thio-U, 5-propynyl-C, LNA-A), because the polymerase step is omitted. Chemical routes thus provide broader immune-silencing palettes but a separate GMP campaign is needed for each modified amidite, while enzymatic routes permit the physical blending of triphosphates in a single pot.
Table 3 Monomer choice dictates mRNA performance
| Monomer | Purpose | Route Limitation | Clinical Example |
| Uridine | Native | Immune-activating | Early research |
| m¹Ψ | Immune silence | Enzymatic + chemical | COVID-19 vaccine |
| 5-methyl-C | CpG depletion | Both routes | All modern mRNA |
| 5-propynyl-C | ↑ Affinity | Chemical only | Research stage |
Nucleoside monomers are the common language of oligonucleotide therapeutics, but the monomer alphabet is widely variant among siRNA, mRNA, antisense and aptamer therapeutics. Although all platforms require GMP-grade, stereochemically pure building blocks, the type, extent and cost sensitivity of chemical modification are all regulated by the therapeutic target pharmacology: siRNA needs high affinity 2'-F caps and flanks, mRNA requires immune silent Ψ or m1Ψ, antisense needs phosphorothioate DNA for RNase H and aptamers use ultra-high affinity LNA or MOE ribosides. This translates into a need for the same solid-phase platform to accept chemically distinct libraries of monomers at a single-base resolution that are precisely targeted for affinity, stability, immunogenicity, and/or nuclease resistance.
GMP-grade monomers are all built on the same floor: ≥99.5 % chemical purity, ≥99.5 % chiral purity, ≤50 ppm water, and ≤5 ppm total metals. But the acceptance criteria for impurities differs between modalities: siRNA can accommodate trace levels of 2'-F epimers, mRNA allows no more than 0.1 % of any Ψ diastereomer, and phosphorothioate DNA must demonstrate control of Sp/Rp isomer drift to <0.5 % over time. As a result, each modality requires its own analytical cascade: chiral LC for Ψ, ion-pair LC-MS for PS stereoisomers, and GC-FID for residual solvents, each fully validated over three consecutive lots to show batch-to-batch reproducibility.
siRNA's use of 2'-F and 2'-O-MOE ribosides with 100 % substitution in guide/passenger strands encodes higher Tm and TLR7/8 silencing. mRNA's use of Ψ or m1Ψ at 100 % uridine substitution encodes avoidance of PKR and RIG-I while maintaining codon decoding. Antisense gapmers' use of phosphorothioate DNA at 100 % backbone substitution encodes RNase H recruitment and is flanked by 2'-O-MOE ribosides at 100 % substitution to provide protection from exonuclease degradation. Aptamers' use of LNA or MOE at selected (30–70 %) positions encodes picomolar affinity while still maintaining flexibility. Modification level is encoded at the monomer stage and is not post-synthetically applied.
mRNA needs kilograms of NTPs, enzymatic polymerisation is single step so cost is set by triphosphate cost not number of cycles. ASO gapmers also need multi-kilogram amounts of amidites, but the 20-mer length means that the mass per oligo is manageable; phosphorothioation adds a sulfur expense, but again the short length limits the overall expense. siRNA is a double-stranded format, so it has twice the monomer demand. 2'-F and 2'-O-Me are specialty amidites that are 3–5× more expensive than DNA, so cost-per-dose is sensitive to the number of modifications. Aptamers are short (15–40 nt), but heavily modified so unit cost is high but mass per patient is low, a combination that is well matched with niche indications. Since scale-up is feedstock limited, multi-purpose plants change amidite bottles instead of reconfiguring reactors, coupling modality flexibility with capital efficiency.
We provide a comprehensive range of nucleoside monomer solutions designed to support the unique chemical and performance requirements of siRNA, mRNA, and antisense oligonucleotide (ASO) therapeutics. By combining high-quality raw materials with deep technical expertise, we help developers achieve consistent synthesis performance, optimized drug properties, and scalable manufacturing across diverse oligonucleotide platforms.
Our portfolio includes a full spectrum of native and modified nucleoside monomers covering all canonical bases required for RNA and DNA therapeutics. This includes ribose- and deoxyribose-based nucleosides, as well as synthesis-ready derivatives optimized for downstream conversion into phosphoramidites or other activated intermediates. All monomers are manufactured to stringent quality standards, ensuring high purity, controlled impurity profiles, and batch-to-batch consistency. This reliability is critical for maintaining coupling efficiency, sequence fidelity, and reproducible performance in oligonucleotide synthesis.
Each oligonucleotide modality presents distinct design and manufacturing challenges. We provide platform-specific nucleoside monomer solutions tailored to the requirements of siRNA, ASO, and mRNA development. For siRNA, our monomers support precise modification patterns that enhance stability, minimize off-target effects, and preserve RNA interference activity. For ASOs, we offer nucleoside derivatives compatible with both RNase H–dependent and steric-blocking mechanisms, enabling optimized binding affinity and pharmacokinetic profiles. For mRNA, we supply nucleosides suitable for both chemical and enzymatic synthesis approaches, supporting efficient translation and controlled immunogenicity.
Selecting the right nucleoside monomers is a critical factor in successful oligonucleotide development. Our technical team provides expert support for monomer selection and optimization, helping customers align raw material choices with therapeutic goals, synthesis strategies, and scale-up plans. We assist with evaluating modification options, protecting group compatibility, and process robustness to ensure that monomers perform reliably throughout development and manufacturing. This collaborative approach helps reduce technical risk and supports efficient program progression.
We support oligonucleotide programs from early discovery through GMP manufacturing with flexible and scalable supply options. From small research quantities to larger-volume clinical and commercial supply, our nucleoside monomers are supported by clear specifications, traceability, and appropriate documentation. This end-to-end supply capability enables smooth transitions between development stages while maintaining consistent quality and regulatory readiness as program requirements evolve.
If you are developing siRNA, mRNA, or antisense oligonucleotide therapeutics, our team is ready to support your program with high-quality nucleoside monomers and expert technical guidance. Contact us today to request technical information, samples, or quotations, or to discuss customized monomer solutions tailored to your RNA or ASO development needs.
References
Yes, they are essential for siRNA, ASO, and mRNA platforms.
Yes, each modality has unique modification and purity requirements.
They influence stability, binding affinity, and synthesis efficiency.
Yes, modification patterns vary by therapeutic goal.
Yes, with a comprehensive and flexible monomer portfolio.

